Validating Vitellogenin Gene Functions in Fertility: From Molecular Mechanisms to Biomedical Applications
This comprehensive review synthesizes current research on vitellogenin (Vtg) gene functions in fertility across diverse species.
Validating Vitellogenin Gene Functions in Fertility: From Molecular Mechanisms to Biomedical Applications
Abstract
This comprehensive review synthesizes current research on vitellogenin (Vtg) gene functions in fertility across diverse species. We explore the foundational biology of Vtg and its receptor (VgR), detailing their crucial roles in yolk formation and oocyte development. The article examines methodological approaches for functional validation, including RNA interference, CRISPR-based techniques, and standardized biomarker assays. We address key challenges in troubleshooting experimental variability and optimizing protocols for reliable results. Finally, we present comparative validation evidence from aquatic organisms, insects, amphibians, and nematodes, highlighting conserved reproductive mechanisms and their implications for developing novel fertility interventions and environmental assessments. This resource provides researchers and drug development professionals with an integrated perspective on Vtg as both a fundamental reproductive protein and a valuable biomarker.
The Essential Biology of Vitellogenin: Molecular Structure and Evolutionary Conservation in Reproduction
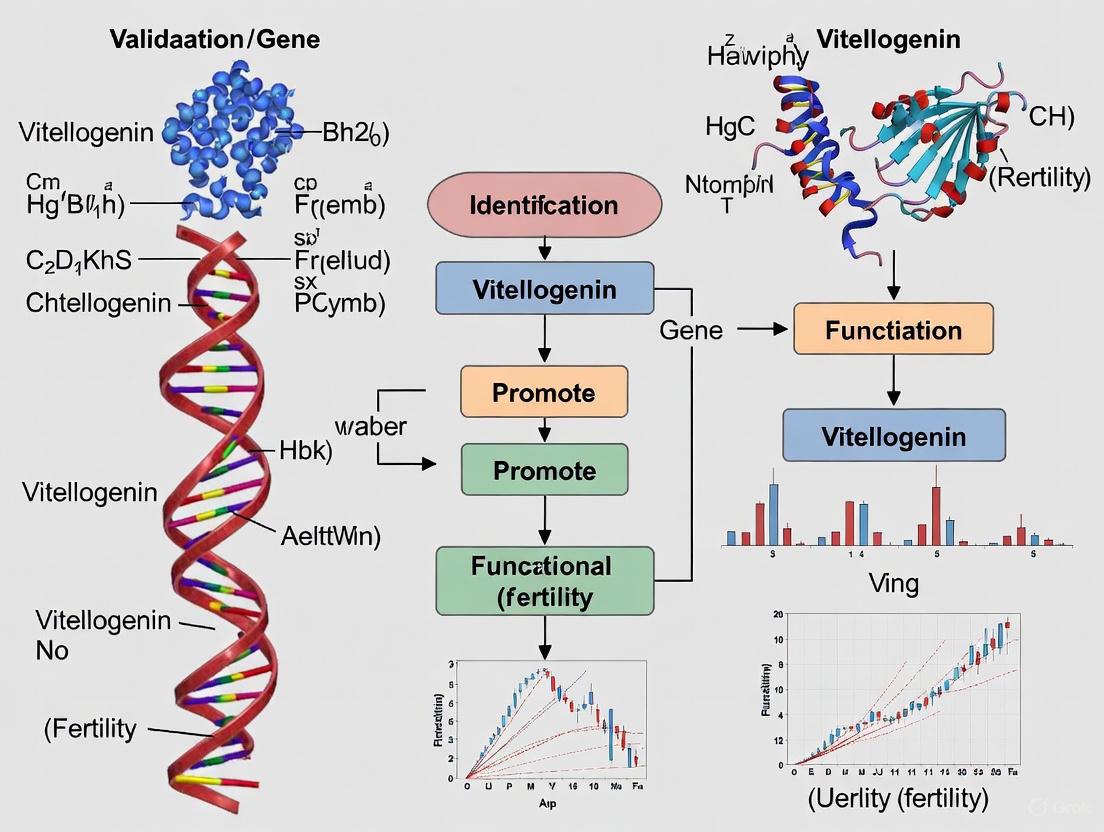
Vitellogenin (Vtg) is a complex lipoglycophosphoprotein that serves as the primary precursor to egg yolk proteins in nearly all oviparous species [1] [2]. This multifunctional molecule belongs to the large lipid transfer protein (LLTP) superfamily, which also includes apolipoprotein B (apoB-100) and the microsomal triglyceride transfer protein (MTP) [3] [1] [2]. The Vtg molecule is synthesized in somatic tissues—typically the liver of vertebrates or the fat body of insects—and is transported via circulation to developing oocytes, where it is internalized via receptor-mediated endocytosis to provide essential nutrients for embryonic development [1] [4] [5]. The structural complexity of Vtg, characterized by its distinctive domains and conserved motifs, underpins its diverse functions in reproduction, lipid transport, and beyond. This review comprehensively compares Vtg's structural domains and functional motifs across species, providing researchers with experimental frameworks for investigating its role in fertility.
Structural Domain Architecture of Vitellogenin
The vitellogenin protein exhibits a conserved multi-domain architecture that facilitates its role as a nutrient transport vehicle. Bioinformatics and structural biology studies have revealed consistent organizational patterns across diverse taxa.
Conserved Domain Organization
A complete vitellogenin polypeptide is composed of several structurally and functionally distinct domains. From N- to C-terminus, these include: a signal peptide for cellular export, lipovitellin-1 (LvH), a phosvitin (Pv) domain, lipovitellin-2 (LvL), and a von Willebrand factor type D (vWD) domain (YGP40) [2] [6]. The recently solved cryo-EM structure of honey bee (Apis cerana) Vtg at 3.20 Ã… resolution further identified a C-terminal cystine knot domain, previously classified as a domain of unknown function [7]. This structural elucidation provides unprecedented insight into the spatial arrangement of these domains and their functional interfaces.
The N-terminal region of Vtg shows significant structural homology to the lipid-binding domains of MTP and apoB, forming a globular β-sheet structure that facilitates lipid recruitment and transport [3] [2]. This region contains the vital vitellogenin_N domain (PF01347), which represents a conserved structural motif found in several lipid transport proteins [2]. Molecular modeling studies indicate that conserved structural motifs forming reciprocal homodimerization interfaces in Vtg are repurposed in MTP to form stable heterodimers with protein disulfide isomerase (PDI), highlighting the evolutionary conservation of these structural elements [3].
Molecular Characteristics and Motif Conservation
Analysis of Vtg sequences across multiple species reveals conserved motif patterns despite sequence divergence. A comprehensive motif analysis of Vtg proteins from 15 vertebrate species identified three conserved motifs present across all sequences, though their starting positions varied between species [6]. These motifs exhibited exceptionally low E-values (4.6e-649, 8.3e-520, and 1.5e-494 respectively), indicating high statistical significance and conservation [6].
In Caenorhabditis elegans, the six vitellogenin genes encode polypeptides that associate into two distinct oligomeric lipoprotein complexes. The B complex is a simple dimer of YP170B, while the A complex is an oligomer composed of YP170A, YP115, and YP88, with the latter two deriving from proteolytic cleavage of VIT-6 [4]. These complexes have estimated molecular weights of approximately 437-439 kDa and diameters of 12.8-14.6 nm [4].
Table 1: Conserved Vitellogenin Domains and Their Functional Roles
| Domain | Structural Features | Primary Functions | Conservation Across Species |
|---|---|---|---|
| Vitellogenin_N | β-sheet rich lipid transport domain | Lipid binding and transport, structural homology to MTP/apoB | High - present in all Vtg and related lipid transport proteins [3] [2] |
| Lipovitellin-1 | Large hydrophobic binding cavities | Lipid sequestration, nutrient storage | High - cleaved from all vertebrate Vtg precursors [2] [6] |
| Phosvitin | Serine-rich, highly phosphorylated | Mineral binding (Ca, Fe), phosphorus storage | Variable - size and phosphorylation degree varies [4] [6] |
| Lipovitellin-2 | Smaller lipid-binding domains | Additional lipid transport capacity | High - consistently present in Vtg proteins [2] [6] |
| vWF type D | Conserved cysteine residues | Structural stabilization, potential protein interactions | Moderate - identified in multiple but not all Vtg types [7] [6] |
| C-terminal CTCK | Cystine knot formation | Structural integrity, potential signaling functions | Recent discovery in honey bee Vtg [7] |
The phosvitin domain deserves special emphasis for its unique biochemical properties. This serine-rich region undergoes extensive phosphorylation, creating a highly anionic domain that chelates calcium, iron, and other cations [4] [6]. In chicken Vtg, phosvitin contains up to 116 mol of phosphorus per mole of protein, accounting for its exceptional metal-binding capacity [6]. The degree of phosphorylation and precise organization of phosvitin domains varies between Vtg types, even within the same species, suggesting functional specialization [6].
Comparative Functional Analysis Across Species
Vitellogenin's structural conservation enables its fundamental role in reproduction, while species-specific variations support specialized biological functions beyond nutrient provision.
Reproduction and Nutrient Provision
The primary function of Vtg across all oviparous species is to provision the developing oocyte with essential nutrients. Vtg serves as a carrier for lipids, carbohydrates, phosphorous, and metals (including Mg, Ca, and Zn) [1] [4]. In C. elegans, yolk complexes purified from embryos contain approximately 15% lipid by weight, with phospholipids (phosphatidylcholine and phosphatidylethanolamine) comprising over 50% of the total lipid content [4]. Neutral lipids constitute around 30%, with free fatty acids and minimal cholesterol completing the profile [4].
The uptake of Vtg into oocytes occurs via receptor-mediated endocytosis through members of the low-density lipoprotein receptor (LDLR) family [5]. In flathead mullet (Mugil cephalus), two putative vitellogenin receptors belonging to the Lr8/VLDLR and Lrp13/LRX + 1 subfamilies have been identified, exhibiting ovary-specific expression profiles consistent with their role in Vtg uptake during oocyte development [5]. These receptors feature conserved ligand-binding domains that facilitate efficient endocytosis of yolk precursors.
Non-Traditional Functions in Social Insects and Beyond
Research in social insects, particularly honey bees (Apis mellifera), has revealed remarkable functional diversification of Vtg. In these species, Vtg has acquired roles in social behavior, longevity, immunity, and antioxidant protection [1] [7]. Honey bee Vtg extends queen and forager lifespan by acting as an antioxidant, with higher Vtg titers in nurse bees correlating with delayed foraging onset and preferential pollen collection [1] [2].
The molecular basis for these pleiotropic functions is becoming clearer with recent structural insights. The cryo-EM structure of honey bee Vtg reveals a lipophilic cavity within the α-helical domain that may facilitate binding to various ligands, potentially explaining Vtg's anti-inflammatory and immunomodulatory functions [1] [7]. Additionally, Vtg participates in a regulatory feedback loop with juvenile hormone, mutually suppressing each other to influence behavioral development and swarming behavior [2].
Table 2: Functional Comparison of Vitellogenin Across Model Organisms
| Species | Primary Reproductive Function | Additional Physiological Roles | Regulatory Mechanisms |
|---|---|---|---|
| Chicken (Gallus gallus) | Precursor to egg yolk proteins (lipovitellins, phosvitin) [6] | Limited evidence of non-reproductive functions | Estrogen-dependent expression in liver [6] |
| Honey Bee (Apis mellifera) | Yolk provision in queens [1] [7] | Longevity, antioxidant activity, immunomodulation, social behavior regulation [1] [7] | Juvenile hormone feedback loop; nutrition-sensitive [1] [2] |
| Nematode (C. elegans) | Nutrient provision for embryogenesis [4] | Supports post-embryonic development; intergenerational signaling [4] | Intestinal expression in hermaphrodites; influenced by environmental cues [4] |
| Fish Models (e.g., Cunner, Zebrafish) | Yolk precursor for oocyte development [8] [9] | Biomarker for estrogenic exposure; potential immune functions [8] [9] | Strong estrogen induction; aromatase regulation [8] [9] |
In C. elegans, Vtg serves not only embryonic development but also supports post-embryonic development and fertility, particularly in harsh environments [4]. Increasing Vtg provisioning underlies several post-embryonic phenotypic alterations associated with advancing maternal age, demonstrating that vitellogenins can act as an intergenerational signal mediating the influence of parental physiology on progeny [4].
Experimental Approaches for Vitellogenin Analysis
Methodologies for Structural and Expression Analysis
Researchers have developed specialized protocols for characterizing Vtg structure and function across species. The structural elucidation of honey bee Vtg employed single-particle cryo-electron microscopy at 3.20 Ã… resolution, with the protein one-step purified directly from hemolymph [7]. This approach preserved native post-translational modifications and lipid associations, providing unprecedented insights into the functional architecture of Vtg.
For motif analysis, bioinformatics tools like MEME (Multiple EM for Motif Elicitation) with the OOPS model have proven effective for identifying conserved patterns in Vtg sequences [6]. This method analyzes amino acid sequences in FASTA format, predicting statistically significant motifs (widths between 6-50 amino acids) based on low E-values, with outputs providing graphical alignments and sequence logos representing position-specific probability matrices [6].
Gene expression studies in insects like Apolygus lucorum have utilized quantitative PCR and protein analysis to correlate AlVg expression with fertility parameters [10]. These studies demonstrated significant linear regression correlations between AlVg expression and key population proliferation parameters including nymph mortality rates, female lifespan, fecundity, and egg hatching rates [10].
Vitellogenin as an Ecotoxicological Biomarker
The estrogen-responsive nature of Vtg makes it a valuable biomarker for endocrine disruption in aquatic environments. Male fish typically lack detectable Vtg, but exposure to estrogenic endocrine-disrupting chemicals (EDCs) induces strong Vtg expression [8] [9]. Experimental exposures in cunner (Tautogolabrus adspersus) have shown that male fish exposed to 17β-estradiol, ethynylestradiol, or estrone can produce Vtg at concentrations exceeding 300 mg/mL while maintaining motile sperm production [8].
Standardized testing protocols now incorporate Vtg measurements alongside other endocrine endpoints. Integrated assessment frameworks combine Vtg data with transcriptomic, epigenetic, and histological analyses within Adverse Outcome Pathway (AOP) and weight-of-evidence (WoE) frameworks to provide mechanistic links between receptor activation and reproductive impairment [9].
The Scientist's Toolkit: Essential Research Reagents and Methods
Table 3: Essential Research Reagents and Experimental Tools for Vitellogenin Studies
| Reagent/Resource | Specific Application | Research Utility | Example Implementation |
|---|---|---|---|
| Cryo-EM Infrastructure | High-resolution structure determination | Elucidating domain architecture and lipid binding cavities [7] | Native Vtg purification from hemolymph followed by single-particle analysis [7] |
| MEME Suite Software | Conserved motif identification | Detecting statistically significant patterns in Vtg sequences [6] | OOPS model analysis of multiple Vtg amino acid sequences in FASTA format [6] |
| LDLR Family Antibodies | Receptor binding studies | Characterizing Vtg uptake mechanisms in oocytes [5] | Immunodetection of Lr8/VLDLR and Lrp13 receptors in ovarian tissues [5] |
| Species-Specific Vtg ELISA | Protein quantification | Biomarker assessment in ecotoxicology and reproductive studies [8] [9] | Measuring Vtg induction in male fish plasma after estrogenic exposure [8] |
| qPCR Primers for Vtg Genes | Transcript level analysis | Gene expression profiling in different tissues and conditions [10] | Correlation of AlVg expression with fertility parameters in insects [10] |
| RNAi Constructs | Functional gene validation | Determining Vtg requirement in reproduction and other functions [1] [4] | Vtg knockdown to assess impacts on longevity and behavior in honey bees [1] |
| Geranylgeraniol-d5 (major) | Geranylgeraniol-d5 (major), MF:C20H34O, MW:295.5 g/mol | Chemical Reagent | Bench Chemicals |
| SN-38 4-Deoxy-glucuronide | SN-38 4-Deoxy-glucuronide | SN-38 4-Deoxy-glucuronide for research. Study irinotecan metabolism and glucuronidation. This product is for Research Use Only (RUO). Not for human or veterinary use. | Bench Chemicals |
Vitellogenin represents a remarkable example of evolutionary conservation coupled with functional diversification. Its conserved structural domains—particularly the vitellogenin_N lipid transport domain, the serine-rich phosvitin region, and the newly characterized C-terminal cystine knot domain—provide the structural basis for its essential role in reproduction across oviparous species [7] [2] [6]. The experimental methodologies summarized here, from cryo-EM structural analysis to expression profiling and functional validation, provide researchers with robust tools for investigating Vtg function in fertility research.
The multifunctionality of Vtg, especially evident in social insects where it influences longevity, behavior, and immunity, highlights the potential for discovering novel functions in other species [1] [7]. Furthermore, the sensitivity of Vtg expression to hormonal manipulation and environmental contaminants makes it an invaluable biomarker for reproductive toxicology and environmental assessment [8] [9]. As structural biology techniques advance and multi-omics approaches become more accessible, our understanding of Vtg's structure-function relationships will continue to deepen, potentially revealing new avenues for fertility management and toxicological assessment across species.
Vitellogenin (Vtg) is a glycolipophosphoprotein that serves as the primary precursor of egg yolk, an essential nutrient reserve for embryonic development in egg-laying species [11] [12]. This protein is synthesized in the liver of vertebrates or the fat body of insects, released into the bloodstream (or hemolymph), and incorporated into developing oocytes through receptor-mediated endocytosis [12] [13]. The vitellogenin gene family exhibits remarkable evolutionary plasticity, with gene number varying significantly across taxonomic lineages due to whole genome duplication events and lineage-specific adaptations [11] [14]. This article provides a comprehensive comparison of Vtg gene structure, function, and regulation across invertebrate and vertebrate systems, framed within the context of validating Vtg functions in fertility research. Understanding the evolutionary history and functional diversification of Vtg genes offers valuable insights for developing novel approaches to manage fertility and reproductive disorders.
Evolutionary Origins and Gene Family Expansion
Gene Duplication and Lineage-Specific Adaptations
The vitellogenin gene family has expanded from two ancestral genes present at the beginning of vertebrate radiation through multiple independent duplication events across diverse lineages [11] [12]. Genomic analyses reveal that the vertebrate Vtg gene cluster originated prior to the separation of Sarcopterygii (tetrapod branch) from Actinopterygii (fish branch) over 450 million years ago, a period associated with the second round of whole genome duplication (WGD) [14]. Following WGD events, many duplicated Vtg genes were lost, while others underwent neofunctionalization, acquiring specialized roles in different taxonomic groups [14].
Table 1: Vitellogenin Gene Complement Across Vertebrate Lineages
| Taxonomic Group | Species Example | Vtg Gene Count | Vtg Types Present | Genomic Organization |
|---|---|---|---|---|
| Jawless Fishes | Silver Lamprey (Ichthyomyzon unicuspis) | 1 | Single form | Single gene |
| Chondrichthyans | Catshark (Scyliorhinus torazame) | 1 | Single form | Single gene |
| Non-Teleost Fishes | Spotted Gar (Lepisosteus oculatus) | 3 | Multiple forms | Cluster organization |
| Basal Teleosts | Atlantic Herring (Clupea harengus) | 3 | VtgAa, VtgAb, VtgC | Cluster organization |
| Acanthomorph Teleosts | Medaka (Oryzias latipes) | 4 | VtgAa1, VtgAa2, VtgAb, VtgC | Cluster with lineage-specific duplication |
| Salmonids | Atlantic Salmon (Salmo salar) | 3 | VtgAsa1, VtgAsb, VtgC | Additional duplication from Ss4R WGD |
| Birds | Chicken (Gallus gallus) | 3 | VtgI, VtgII, VtgIII | Conserved cluster on chromosome 8 |
| Egg-Laying Mammals | Platypus (Ornithorhynchus anatinus) | Vtg fragments detected | Residual Vtg genes | Traces of conserved Vtg cluster |
In invertebrates, Vtg genes exhibit different evolutionary patterns. In insects, Vg is typically encoded by a single gene, though it has evolved pleiotropic functions in highly eusocial species like the honey bee (Apis mellifera), where it influences social behavior, foraging specialization, and longevity in addition to its reproductive role [15]. In crustaceans such as the Pacific white shrimp (Litopenaeus vannamei), Vtg gene expression is regulated by members of the crustacean hyperglycemic hormone family, including molt-inhibiting hormone [16].
Structural Evolution of Vtg Proteins
Vitellogenin proteins across taxa share a common multi-domain architecture but exhibit significant structural variations that reflect functional adaptations:
- Vertebrate VtgA-type: Complete proteins containing heavy chain lipovitellin (LvH), phosvitin (Pv), light chain lipovitellin (LvL), von Willebrand factor type D domain (Vwfd) with a beta component (β'), and a C-terminal coding region (CT) [12]
- Vertebrate VtgC-type: Phosvitinless forms lacking the Pv domain, existing primarily as a complex of LvH and LvL [12] [14]
- Insect Vtg: Contains conserved domains similar to vertebrate Vtgs but may have additional functional regions that enable pleiotropic roles in social species [17] [15]
The structural diversity of Vtg genes and proteins across species has important implications for their function in reproduction and other biological processes.
Comparative Analysis of Vtg Gene Functions
Vertebrate Vtg Gene Specialization
In vertebrates, different Vtg paralogs have evolved specialized functions, particularly in teleost fishes where they determine fundamental egg characteristics:
- VtgAa: Generates free amino acids through catheptic proteolysis during oocyte maturation, driving water influx and production of pelagic eggs that float in the marine environment [14]
- VtgAb: Remains largely intact during oocyte maturation, contributes primarily to nutrient provision, and is predominant in species with benthic eggs that sink [14]
- VtgC: Phosvitinless form that provides supplemental nutrition but contributes minimally to oocyte hydration [14]
This functional specialization represents a classic case of neofunctionalization following gene duplication, where duplicated genes evolve new functions rather than simply partitioning ancestral functions.
Invertebrate Vtg Gene Pleiotropy
In invertebrates, Vtg has acquired diverse non-reproductive functions, particularly in eusocial insects:
- Honey Bees: Vitellogenin influences temporal division of labor, foraging specialization (pollen vs. nectar preference), and longevity in worker bees [15]
- Tomato Leaf Miner (Tuta absoluta): Vtg is essential for oocyte development, yolk deposition, and overall female fecundity [17]
The expansion of Vtg functions in invertebrates, especially social insects, demonstrates how reproductive proteins can be co-opted for novel regulatory roles in complex social behaviors.
Table 2: Functional Roles of Vitellogenin Across Taxa
| Organismal Group | Primary Reproductive Function | Non-Reproductive Functions | Specialized Adaptations |
|---|---|---|---|
| Teleost Fishes | Yolk precursor for embryonic nutrition | Limited non-reproductive functions | VtgAa neofunctionalization for pelagic egg production |
| Birds & Reptiles | Yolk precursor for embryonic nutrition | Limited non-reproductive functions | Multiple Vtg forms with differential utilization |
| Egg-Laying Mammals | Yolk precursor (platypus) | Lost in placental mammals | Progressive gene loss in therian mammals |
| Insects | Yolk precursor for oocyte development | Social behavior, longevity, antioxidant protection (honey bee) | Pleiotropic expansion in eusocial species |
| Crustaceans | Yolk precursor for oocyte development | Molt cycle regulation | Regulation by CHH-family neuropeptides |
Experimental Analysis of Vtg Gene Function
Key Methodologies in Vtg Research
Gene Expression Manipulation
- RNA Interference (RNAi): successfully applied in both insects and fish to knock down Vtg gene expression [17] [15]
- Transcriptomic Analysis: RNA sequencing used to identify stage-specific changes in Vtg and related pathway expression [18]
Promoter Analysis
- Reporter Assays: Identification of estrogen-responsive elements in the Vtg receptor promoter [13]
- Electrophoretic Mobility Shift Assays (EMSA): Confirmation of transcription factor binding to Vtg regulatory regions [13]
Structural Biology
- Cryo-Electron Microscopy: Determination of full-length honey bee Vtg structure at 3.2 Ã… resolution [7]
Vitellogenin Uptake Mechanism Investigation
Research in the New Zealand shortfinned eel (Anguilla australis) has employed comparative transcriptomics of pre-vitellogenic and early vitellogenic ovaries to investigate the regulation of Vtg uptake [18]. This approach tested two competing hypotheses regarding the control of Vtg incorporation into oocytes:
Diagram 1: Experimental framework for investigating Vtg uptake mechanisms in eel ovaries. The mechanical barrier hypothesis proposes tight junctions regulate Vtg access, while the chemical barrier hypothesis involves gap junction signaling.
The study provided partial support for the mechanical barrier hypothesis, with seven genes encoding tight junction proteins showing differential expression between pre-vitellogenic and early vitellogenic ovaries [18]. In contrast, the chemical barrier hypothesis was not supported, as gap junction gene expression remained unchanged. Additionally, the research identified components of the endocytic pathway that were upregulated during the transition to vitellogenesis, including putative Vtg receptors corresponding to Lr8 and Lrp13 members of the low-density lipoprotein receptor family [18].
Research Reagent Solutions for Vtg Studies
Table 3: Essential Research Reagents for Vitellogenin Gene Functional Validation
| Reagent/Category | Specific Examples | Research Application | Key Function in Vtg Studies |
|---|---|---|---|
| Gene Expression Manipulation | dsRNA for RNAi (Vtg-specific) | Functional validation through gene knockdown [17] [15] | Reduces Vtg mRNA and protein to assess phenotypic effects |
| Hormone treatments (E2, EE2) | Regulation of Vtg and VtgR expression [13] | Modulates estrogen-responsive Vtg synthesis and uptake | |
| Promoter Analysis Tools | Reporter gene constructs (Luciferase, GFP) | Promoter activation assays [13] | Measures transcriptional activity of Vtg regulatory regions |
| GenomeWalker kits | Promoter isolation [13] | Clones 5' regulatory regions of Vtg and VtgR genes | |
| Structural Biology Reagents | Cryo-EM reagents | Structural determination of native Vtg [7] | Elucidates domain architecture and lipid binding sites |
| Protein purification systems (IMAC) | Recombinant protein production [16] | Generates functional Vtg and regulatory proteins for assays | |
| Transcriptomic Analysis | RNA sequencing kits | Gene expression profiling [18] | Identifies differentially expressed genes in Vtg pathways |
| RNA extraction kits (NucleoSpin) | RNA isolation from tissues [18] | Obtains high-quality RNA for expression studies |
The evolutionary history of vitellogenin genes reveals a complex tapestry of gene duplication, functional specialization, and in some cases, gene loss corresponding to reproductive strategies. From the conserved Vtg cluster in oviparous vertebrates to the pleiotropic expansions in social insects, this gene family demonstrates remarkable adaptability. The experimental methodologies summarized here provide a toolkit for researchers investigating Vtg functions in fertility and reproduction.
For drug development professionals, Vtg genes and their receptors represent potential targets for fertility management. The differential regulation of Vtg paralogs, the stage-specific expression of Vtg receptors, and the endocrine control of Vtg synthesis offer multiple intervention points. Furthermore, the conservation of Vtg genes across taxa facilitates translational approaches, allowing insights from model organisms to inform therapeutic development for human reproductive health.
Future research should focus on elucidating the precise molecular mechanisms through which different Vtg paralogs contribute to oocyte quality, how Vtg receptors are regulated during follicular development, and how environmental factors disrupt Vtg pathways. Such investigations will continue to enhance our understanding of reproductive biology and provide novel strategies for addressing fertility challenges.
The Vitellogenin Receptor (VgR/VLDLR) and Oocyte Endocytosis Machinery
The Vitellogenin Receptor (VgR), also known as the Very Low-Density Lipoprotein Receptor (VLDLR), serves as the principal gatekeeper for yolk deposition in oviparous vertebrates and invertebrates. This receptor mediates the massive uptake of vitellogenin (Vtg)—the precursor of egg yolk proteins—from the maternal circulation into developing oocytes through receptor-mediated endocytosis [19] [2]. The efficiency of this process directly determines embryonic nutritional reserves and consequently impacts fertility outcomes. Within fertility research, characterizing the VgR/VLDLR machinery provides crucial insights into reproductive failures and offers potential biomarkers for assessing fertility status across species [20].
The VgR/VLDLR belongs to the low-density lipoprotein receptor (LDLR) superfamily, which comprises 15 recognized receptors including LDLR, VLDLR, SORLA, and various LDL receptor-related proteins (LRPs) [19]. These receptors share structural similarities but have diversified in function through evolution, with VgR/VLDLR specializing in reproductive nutrient transport. Understanding the molecular architecture and operational mechanisms of this receptor system is fundamental for developing targeted approaches to modulate fertility in both agricultural and biomedical contexts.
Comparative Analysis of VgR/VLDLR Across Species
Structural Organization and Functional Domains
The VgR/VLDLR exhibits a conserved domain architecture across species while possessing distinctive features that reflect evolutionary adaptations. As a member of the LDLR superfamily, it is a type I transmembrane receptor characterized by an extracellular domain containing cysteine-rich ligand-binding repeats (LBRs), a transmembrane domain, and an intracellular domain with conserved signaling motifs [19]. The extracellular domain is responsible for recognizing and binding vitellogenin, while the intracellular domain facilitates clathrin-coated vesicle formation through its Asn-Pro-X-Tyr (NPxY) motif [19].
Table 1: Comparative Structural Features of VgR/VLDLR Across Species
| Species | Receptor Designation | Ligand-Binding Repeats | Conserved Motifs | Distinguishing Features |
|---|---|---|---|---|
| Cynops orientalis (Chinese fire-bellied newt) | VTGR/VLDLR | 8 LDLR class A domains [19] | NPxY cytoplasmic motif [19] | O-glycosylated domain near transmembrane region [19] |
| Bemisia tabaci (whitefly) | BtA1VgR | 12 LDLa, 10 LDLb domains [20] | - | 7 EGF domains, insect-specific adaptations [20] |
| Vertebrates (general) | VLDLR | 8 cysteine-rich repeats [21] | NPxY, O-linked sugar domain [19] | Similar to LDLR but with extra ligand-binding repeat [19] |
| Insects (general) | VgR | Two clusters (5+7) of class A repeats [21] | - | Unique two-cluster organization distinct from vertebrate receptors [21] |
Comparative analyses reveal that while the fundamental blueprint of VgR/VLDLR is conserved, specific structural variations have emerged through evolution. In the amphibian Cynops orientalis, VTGR shares the characteristic eight-module ligand-binding domain organization with other vertebrate VLDLRs, but also contains specific O-glycosylation sites that may influence receptor function [19]. Insect VgRs, such as those characterized in Bemisia tabaci, display a distinctive two-cluster organization of ligand-binding repeats that phylogenetic evidence suggests arose independently from the single-cluster vertebrate configuration [20] [21]. These structural differences reflect divergent evolutionary paths while maintaining the core function of vitellogenin uptake.
Expression Patterns and Functional Validation
The expression profile and functional role of VgR/VLDLR have been systematically characterized across multiple species, revealing both conserved and species-specific aspects of its operation in oogenesis.
Table 2: Expression Profiles and Functional Validation of VgR/VLDLR
| Species | Tissue Expression | Functional Role | Validation Method | Key Findings |
|---|---|---|---|---|
| Cynops orientalis [19] | Ovarian tissue, hepatic tissue [19] | Vitellogenin uptake in oocytes [19] | Transcriptome analysis, phylogenetic comparison [19] | LRP8 also expressed in ovary, suggesting possible complementary role in vitellogenesis [19] |
| Bemisia tabaci [20] | Oocyte membrane [20] | Vg transport from hemolymph to oocytes [20] | siRNA silencing, fecundity assessment [20] | VgR silencing reduced fecundity without affecting Vg transcript levels [20] |
| Xenopus species [22] | Oocyte membrane [22] | Vg endocytosis and yolk platelet formation [22] | Electron microscopy, subcellular fractionation [22] | Vg traverses tubular endosomes to multivesicular bodies for processing [22] |
| Tribolium castaneum [23] | Oocyte membrane (inferred) | Vg uptake (indirect regulation) [23] | RNAi, hormone manipulation [23] | JH regulates Vg through insulin-like peptide signaling pathway [23] |
Expression analyses consistently demonstrate ovarian-specific localization of VgR/VLDLR across species, underscoring its specialized role in reproduction. In Cynops orientalis, transcriptomic studies revealed robust expression of VTGR in ovarian tissues, supporting its function as the primary vitellogenin receptor, while also detecting LRP8 expression, suggesting potential functional redundancy or complementary roles for multiple LDLR family members in vitellogenesis [19]. Functional validation through receptor silencing in Bemisia tabaci established a direct correlation between VgR expression and reproductive success, with silenced insects exhibiting significantly reduced fecundity despite normal vitellogenin transcription [20]. This confirms the receptor's rate-limiting role in yolk deposition independent of vitellogenin production.
Experimental Approaches for VgR/VLDLR Characterization
Receptor Identification and Phylogenetic Analysis
The characterization of VgR/VLDLR begins with comprehensive identification and evolutionary placement through bioinformatic approaches. A representative protocol from recent research involves:
Step 1: Sequence Retrieval and Assembly
- Extract RNA from hepatic and gonadal tissues
- Construct and sequence cDNA libraries
- Perform de novo transcriptome assembly
- Identify putative VgR/VLDLR sequences through homology searches [19]
Step 2: Phylogenetic Reconstruction
- Compile sequences of all 15 LDLR superfamily members from multiple vertebrate genera
- Employ multiple sequence alignment algorithms
- Construct maximum likelihood or Bayesian inference trees
- Assess bootstrap support for clade reliability [19]
Step 3: Synteny Analysis
- Examine genomic localization of receptor genes
- Identify conserved microsyntenic relationships
- Detect species-specific gene duplications [19]
This integrated approach successfully identified 12 complete LDLR superfamily sequences in Cynops orientalis, with phylogenetic analysis of 161 sequences across 11 vertebrate genera confirming VTGR's position within the conserved VLDLR/LDLR/LRP8 clade [19]. The study also revealed species-specific gene duplication events, such as duplicate copies of lrp1, lrp8, and lrp13 in the pufferfish Takifugu rubripes, suggesting functional diversification in different lineages [19].
Functional Validation Through Receptor Silencing
RNA interference provides a powerful tool for establishing causal relationships between VgR/VLDLR expression and reproductive outcomes. A standardized protocol for functional validation includes:
Step 1: dsRNA/siRNA Design and Synthesis
- Identify unique 300-500 bp target sequence
- Incorporate T7 promoter sequences for in vitro transcription
- Synthesize dsRNA using MEGAscript T7 kit [23]
- Purify and quantify dsRNA products
Step 2: Delivery System
- Anesthetize adult insects with ether vapor
- Microinject dsRNA (400 ng/insect) into abdominal segment
- Use aspirator tube assembly with glass capillary needle [23]
- Include control group receiving non-target dsRNA
Step 3: Phenotypic Assessment
- Monitor mortality daily
- Quantify fecundity (eggs laid per female)
- Analyze Vg accumulation in oocytes via Western blot
- Examine oocyte development histologically [20]
Application of this methodology in Bemisia tabaci demonstrated that BtA1VgR silencing specifically impaired Vg protein accumulation in oocytes without altering Vg transcript levels, establishing the receptor's essential role in Vg transport rather than synthesis [20]. The significant mortality and reduced fecundity observed in silenced insects further confirmed VgR's critical function in reproductive success [20].
Tracing Endocytic Pathways
Understanding the intracellular journey of vitellogenin following receptor binding requires sophisticated tracer methodologies. A classic approach involves:
Step 1: Tracer Preparation
- Adsorb purified vitellogenin to colloidal gold particles
- Characterize Vg-gold conjugate size and stability [22]
Step 2: Experimental Incubation
- Expose isolated oocytes to Vg-gold conjugates
- Utilize pulse-chase design with varying incubation times
- Include controls with excess unlabeled Vg to assess specificity [22]
Step 3: Ultrastructural Analysis
- Process samples for transmission electron microscopy
- Perform subcellular fractionation on sucrose density gradients
- Correlate biochemical fractions with morphological compartments [22]
This experimental paradigm in Xenopus oocytes established the central role of multivesicular bodies (MVBs) as key processing compartments in vitellogenin endocytosis [22]. The research demonstrated that Vg enters through coated pits and vesicles, rapidly appears in tubular endosomes and MVBs, and remains in MVBs for extended periods while undergoing condensation into crystalline yolk platelets [22]. The transformation of MVBs into mature yolk platelets represents the final stage of Vg processing for long-term storage.
The Endocytosis Machinery: Molecular Mechanisms
Vitellogenin Internalization Pathway
The intracellular trafficking of vitellogenin following VgR-mediated endocytosis follows a conserved pathway with distinct organellar transitions:
Diagram 1: Vitellogenin intracellular trafficking pathway. Following receptor binding, Vg progresses through specialized compartments culminating in mature yolk platelets.
The endocytic pathway begins with vitellogenin binding to VgR localized in clathrin-coated pits on the oocyte membrane [22]. Following internalization, the receptor-ligand complex enters the endosomal system, where the acidic environment facilitates dissociation. The vitellogenin then progresses to multivesicular bodies (MVBs), which serve as the primary site for Vg condensation and storage [22]. In MVBs, vitellogenin undergoes progressive processing and crystallization, gradually transforming into definitive yolk platelets that provide nutritional support for embryonic development [22].
The entire process is highly regulated, with the final transition from light MVBs (density ~1.21 g/cc) to heavy yolk platelets (density ~1.23 g/cc) representing the rate-limiting step in Vg transport [22]. This elaborate cellular machinery enables oocytes to accumulate massive yolk reserves efficiently, with a single Xenopus oocyte capable of importing approximately 10¹² vitellogenin molecules during its development [22].
Regulatory Signaling Networks
Vitellogenin receptor expression and function are embedded within complex endocrine signaling networks that coordinate reproductive readiness with nutritional status:
Diagram 2: Regulatory network controlling vitellogenin production and uptake. JH and nutritional signals integrate through insulin signaling pathways.
The regulatory circuitry involves juvenile hormone (JH) acting as the primary gonadotropic signal that stimulates vitellogenin gene expression in the fat body (insects) or liver (vertebrates) [23] [24]. JH functions through the insulin-like peptide signaling pathway, inducing expression of insulin-like peptides (ILPs) that in turn suppress the FOXO transcription factor, thereby relieving its repression of vitellogenin genes [23]. This hierarchical arrangement ensures that vitellogenin production is precisely coordinated with nutritional status, as ILPs serve as sensors of nutrient availability. While direct regulation of VgR expression by JH remains less characterized, the receptor's function is ultimately dependent on this integrated signaling network to ensure synchronized Vg availability and uptake capacity.
Research Reagent Solutions for VgR/VLDLR Studies
Table 3: Essential Research Reagents for VgR/VLDLR Investigations
| Reagent Category | Specific Examples | Research Applications | Key Considerations |
|---|---|---|---|
| Gene Silencing Tools | dsRNA targeting VgR/VLDLR [20] [23], siRNA constructs [20] | Functional validation through RNA interference | Species-specific sequence design; optimal target length 300-500 bp for dsRNA [23] |
| Antibody Reagents | Anti-Vg polyclonal antibodies [23], anti-phospho-AKT [23], anti-FOXO [23] | Western blotting, immunohistochemistry, receptor localization | Validate cross-reactivity for non-model organisms; check phosphorylation status for signaling studies [23] |
| Tracer Compounds | Vitellogenin-gold conjugates [22], ¹²âµI-Vg-Au [22] | Ultrastructural tracing of endocytic pathways | Control for non-specific uptake; optimize particle size for different species [22] |
| Hormone Modulators | Juvenile hormone analogs (methoprene) [23], bovine insulin [23] | Signaling pathway manipulation | Dose-response determination critical; consider timing of administration relative to vitellogenic cycle [23] |
| Molecular Biology Kits | SMARTer RACE cDNA Amplification Kit [20], First Strand cDNA Synthesis Kit [20] | Receptor gene cloning and expression analysis | RNA integrity crucial for full-length transcript amplification; utilize species-specific RACE adapters [20] |
The selection of appropriate research reagents is critical for successful investigation of VgR/VLDLR function. RNA interference reagents must be designed to target unique regions of the receptor sequence to ensure specificity, with proper controls including non-target dsRNA and untreated groups [20] [23]. Antibody reagents enable both localization and quantification studies, but require thorough validation in each experimental system due to potential cross-reactivity issues with related LDLR family members [23]. Tracer compounds like Vg-gold conjugates provide powerful tools for visualizing the intracellular journey of vitellogenin, though researchers must optimize particle size and concentration to avoid disrupting normal physiological processes [22].
The Vitellogenin Receptor and its associated endocytosis machinery represent a sophisticated biological system dedicated to reproductive success. The comparative data presented herein establishes both conserved principles and species-specific adaptations in VgR/VLDLR structure and function. The experimental methodologies and reagent frameworks outlined provide researchers with validated approaches for investigating this critical receptor system in diverse biological contexts.
Future research directions should focus on elucidating the precise regulatory mechanisms controlling VgR gene expression, particularly its integration with nutritional sensing pathways. Additionally, structural characterization of VgR-Vg complexes would advance understanding of ligand-receptor specificity and facilitate development of targeted fertility modulators. The expanding availability of genomic resources across oviparous species will enable more comprehensive evolutionary analyses of VgR/VLDLR diversification and specialization. These research avenues promise to yield important insights with applications ranging from agricultural productivity to conservation of endangered species and understanding the fundamental mechanisms governing fertility.
Vitellogenin (Vtg), the precursor of yolk protein, is a critical determinant of fertility in oviparous species. Its production and uptake are orchestrated by complex hormonal networks that integrate developmental and environmental cues. Understanding these regulatory mechanisms is not only fundamental to reproductive biology but also pivotal for developing novel strategies in pest control and addressing human fertility challenges. This guide provides a comparative analysis of the hormonal control of Vtg across model organisms, synthesizing experimental data to validate Vtg gene functions in fertility research. We objectively compare findings from crustaceans, insects, and nematodes, offering researchers a structured overview of conserved and species-specific regulatory paradigms.
Comparative Hormonal Regulation of Vitellogenin
The regulation of vitellogenin (Vtg) exhibits both conserved principles and remarkable diversity across species. The table below summarizes the key hormonal regulators and their mechanisms of action.
Table 1: Comparative Overview of Hormonal Control of Vtg Expression and Secretion
| Organism/Group | Key Regulatory Hormone(s) | Primary Site of Vg Synthesis | Effect on Vg Expression/Secretion | Experimental Evidence |
|---|---|---|---|---|
| Crustaceans (e.g., Callinectes sapidus) | Molt-Inhibiting Hormone (MIH) | Hepatopancreas | Stage-dependent; stimulates Vtg mRNA and protein secretion at mid-vitellogenesis (ovarian stage 3) [25]. | In vitro hepatopancreas incubation, RIA, QPCR [25]. |
| Insects (e.g., Lasioderma serricorne, Tuta absoluta) | Juvenile Hormone, Ecdysone (inferred) | Fat Body, Ovaries | Critical for ovarian development; RNAi silencing reduces Vg content, fecundity, and egg hatchability [26] [17]. | RNAi-mediated gene silencing, qPCR, phenotypic observation of ovaries and fecundity [26] [17]. |
| Nematodes (e.g., Caenorhabditis elegans) | Insulin/IGF-1 Signaling, Steroid Hormone Signaling | Intestine | Sustains post-embryonic development and fertility; regulated by major signaling pathways and environmental experience [27]. | Mutant analysis, transcriptomics, proteomics [27]. |
| Vertebrates (e.g., Birds, Fish) | Estrogen | Liver | Induces massive Vg synthesis in females; expression can be induced in males with exogenous estrogen [27]. | Hormonal induction, ligand-binding assays [27]. |
Key Regulatory Pathways and Network Motifs
The hormonal control of Vtg is embedded within larger regulatory networks that often feature feedback and feedforward loops to ensure precise control over reproductive timing and resource allocation.
- Stage-Dependent Regulation: In the blue crab (Callinectes sapidus), MIH titers are four times higher at mid-vitellogenesis compared to pre-vitellogenesis, highlighting a stage-specific role [25]. The action of MIH is also stage-dependent, showing a stimulatory effect on Vtg mRNA and secretion at advanced ovarian stages, but a more mixed response at early stages [25].
- Conserved Systemic Regulation: In insects and vertebrates, Vtg production is typically under the control of systemic hormones. While not detailed in the provided insect studies, the well-established roles of juvenile hormone and ecdysteroids are inferred [26] [17]. In vertebrates, estrogen is the primary regulator of Vg gene transcription in the liver [27].
- Intergenerational Signaling: In C. elegans, vitellogenins act as more than just a nutrient source; they function as an intergenerational signal. Changes in vitellogenin provisioning based on parental physiology or environmental experience can directly influence progeny development and phenotype [27].
The following diagram illustrates a generalized, cross-species regulatory network for Vtg expression, integrating key hormonal stimuli and feedback mechanisms.
Figure 1: Generalized hormonal regulation network of Vtg expression and its role in fertility. CNS: Central Nervous System.
Experimental Data and Validation of Vtg Function
The critical role of Vtg and its receptor (VgR) in female reproduction has been robustly validated through targeted gene disruption experiments, primarily using RNA interference (RNAi).
Quantitative Impact of Vtg and VgR Gene Silencing
The functional validation of Vtg and VgR genes consistently demonstrates their non-redundant role in female fertility. The quantitative data from key experiments are summarized below.
Table 2: Phenotypic Consequences of Vtg/VgR Gene Silencing on Female Reproduction
| Species | Target Gene(s) | Effect on Oocyte Length | Effect on Fecundity (Egg Number) | Effect on Egg Hatch Rate | Reference |
|---|---|---|---|---|---|
| Lasioderma serricorne (Cigarette Beetle) | LsVg | Significant decrease | Significantly reduced | Significantly reduced | [26] |
| Lasioderma serricorne (Cigarette Beetle) | LsVgR | Significant decrease | Significantly reduced | Significantly reduced | [26] |
| Lasioderma serricorne (Cigarette Beetle) | LsVg + LsVgR | More pronounced decrease | More significantly reduced | Information not specified | [26] |
| Tuta absoluta (Tomato Leafminer) | TaVg | Shorter ovarian tubes, fewer oocytes | Significantly reduced | Significantly reduced | [17] |
Detailed Experimental Protocols
The consistency of findings across species, as shown in Table 2, is underpinned by rigorous and replicable experimental methodologies.
RNAi-Mediated Gene Silencing and Phenotypic Analysis in Insects
This protocol, adapted from studies on Lasioderma serricorne [26] and Tuta absoluta [17], is a standard for functional gene validation in insect fertility research.
Gene Cloning and dsRNA Synthesis:
- Total RNA is isolated from adult insects (typically whole bodies or specific tissues like the ovary or fat body) using reagents like TransZol.
- The open reading frame (ORF) of the target Vg or VgR gene is amplified from a cDNA template via PCR using gene-specific primers.
- The PCR product is ligated into a cloning vector (e.g., pGEM-T Easy Vector) and sequenced for verification.
- Double-stranded RNA (dsRNA) is synthesized in vitro using a high-yield transcription kit (e.g., TranscriptAid T7) with primers containing T7 promoter sequences. The dsRNA is purified and dissolved in nuclease-free water.
dsRNA Delivery:
- For insects like L. serricorne and T. absoluta, micro-injection is the primary delivery method. A defined quantity (e.g., 200 ng) of dsRNA is injected into the hemocoel of female pupae or early-stage adults. Control groups are injected with dsRNA targeting an unrelated gene (e.g., GFP) or with nuclease-free water.
Efficiency and Efficacy Assessment:
- Molecular Validation: The efficiency of gene knockdown is confirmed using quantitative real-time PCR (qPCR). Total RNA is extracted from treated and control insects, and the relative expression level of the target Vg/VgR gene is calculated using the 2^(-∆∆CT) method, with reference genes like EF1a and 18S for normalization [26].
- Biochemical Validation: The vitellogenin content in the hemolymph or whole body is measured to confirm the functional knockdown at the protein level [26].
Phenotypic Characterization:
- Ovarian Development: Ovaries are dissected from treated and control adults. Key metrics include the length of the ovarian tubes, the size of the basal oocytes, and the overall morphology and yolk deposition in the egg chambers [26] [17].
- Fecundity and Fertility Assays: Treated and control virgin females are mated with untreated males. The total number of eggs laid per female (fecundity) is recorded daily. The egg hatching rate (hatchability) is tracked to assess fertility [26] [17].
The workflow for this functional validation is outlined below.
Figure 2: Experimental workflow for RNAi-mediated validation of Vtg gene function.
In Vitro Hepatopancreas Incubation System in Crustaceans
The study on the blue crab (Callinectes sapidus) utilized an in vitro system to dissect the direct effects of neurohormones on Vtg production [25].
- Tissue Preparation: Hepatopancreas fragments are dissected from females at specific ovarian developmental stages (e.g., early vs. mid-vitellogenesis).
- Incubation and Treatment: The tissue fragments are incubated in a plain culture medium. The stability of total RNA and Vtg mRNA is verified over the incubation period (e.g., 6 hours). Fragments are then treated with physiological doses of hormones like MIH or CHH.
- Molecular Analysis:
- Vtg Secretion: The level of Vtg protein secreted into the culture medium is quantified using a specific radioimmunoassay (RIA).
- Gene Expression: Vtg mRNA levels in the tissue are measured using quantitative PCR (QPCR). Heterogeneous nuclear (hn)VtG RNA can also be measured as an indicator of transcription rates.
- Inhibition Assays: To confirm the mechanism of action, inhibitors like Actinomycin D (a transcription inhibitor) and Cycloheximide (a translation inhibitor) are used in conjunction with hormone treatment. For instance, Actinomycin D blocks the stimulatory effect of MIH on Vtg mRNA and protein, confirming its action is at the transcriptional level [25].
The Scientist's Toolkit: Essential Research Reagents
The following table catalogues key reagents and materials essential for conducting research on Vtg regulation and function, as derived from the cited experimental protocols.
Table 3: Essential Research Reagents for Vitellogenin Functional Studies
| Reagent/Material | Function in Research | Specific Examples from Literature |
|---|---|---|
| Cloning & Vector Systems | Amplification and sequencing of target Vg/VgR genes. | pGEM-T Easy Vector [26]. |
| In Vitro Transcription Kits | Synthesis of high-quality, gene-specific double-stranded RNA (dsRNA) for RNAi. | TranscriptAid T7 High Yield Transcription Kit [26]. |
| Micro-injection Apparatus | Precise delivery of dsRNA or other reagents into the hemocoel of small organisms. | Used for dsRNA delivery in L. serricorne and T. absoluta pupae/adults [26] [17]. |
| RNA Isolation Reagents | Extraction of high-integrity total RNA from tissues for downstream molecular analysis. | TransZol reagent [26]. |
| qPCR SuperMix & Instruments | Quantification of gene expression levels (e.g., knockdown efficiency, Vg mRNA levels). | TransStart Top Green qPCR SuperMix [26]. |
| Specific Radioimmunoassay (RIA) | Precise quantification of vitellogenin protein levels in hemolymph, tissue, or culture medium. | Used to measure VtG secretion in blue crab hepatopancreas incubation system [25]. |
| Transcriptional/Translational Inhibitors | Mechanistic studies to determine the level (transcriptional vs. translational) of hormonal regulation. | Actinomycin D and Cycloheximide used in blue crab study [25]. |
| TMRM Chloride | TMRM Chloride, MF:C25H25ClN2O3, MW:436.9 g/mol | Chemical Reagent |
| 25-Epitorvoside D | 25-Epitorvoside D, MF:C38H62O13, MW:726.9 g/mol | Chemical Reagent |
The comparative analysis of Vtg regulation underscores a universal principle: successful reproduction depends on exquisitely timed hormonal control of vitellogenin synthesis and uptake. While the specific hormonal actors may differ—MIH in crustaceans, juvenile hormone in insects, or estrogen in vertebrates—the output, Vtg production, is a conserved linchpin of fertility. The experimental data, consistently showing that targeted disruption of Vtg or VgR severely impairs oogenesis and fecundity, validates these genes as high-value targets for both agricultural pest management and fundamental reproductive science. The toolkit of methods, particularly RNAi, provides a powerful and transferable approach for functional validation across species. A deeper systems-level understanding of the feedback and feedforward networks governing Vtg expression, especially in response to environmental factors, represents a critical frontier for future research with implications for ecology, agriculture, and medicine.
The vitellogenin (Vtg) gene family, which encodes the major yolk protein precursors in oviparous species, provides a powerful model for studying how gene multiplicity drives functional diversification and species-specific adaptations. In oviparous vertebrates and invertebrates, Vtgs are essential for reproductive success, serving as the primary nutritional source for developing embryos [28] [4]. Beyond this conserved role, recent research has revealed that Vtg genes have undergone lineage-specific expansions, resulting in paralogous genes with specialized functions that correlate with diverse ecological niches and reproductive strategies [29] [30]. This guide systematically compares Vtg gene multiplicity and its functional consequences across key model organisms, providing researchers with experimental data, methodological protocols, and analytical frameworks for investigating Vtg gene function in fertility research.
Vtg Gene Multiplicity Across Species
Gene duplication events, including whole-genome duplications (WGDs) and lineage-specific tandem duplications, have shaped the Vtg gene repertoire across taxa. The table below summarizes the number and types of Vtg genes identified in major model organisms.
Table 1: Vitellogenin Gene Multiplicity Across Species
| Species | Classification | Number of Vtg Genes | Gene Designations | Evolutionary Origin |
|---|---|---|---|---|
| Flathead Mullet (Mugil cephalus) | Teleost Fish | 2 putative Vtg receptors | Lr8/VLDLR, Lrp13/LRX+1 | Characterized from 87 LDLR family members [28] |
| Honeybee (Apis mellifera) | Insect | 1 | - | Co-opted for social behavior regulation [15] |
| Mosquitoes (Aedes, Culex, Anopheles) | Insects | 3-4 | AeVgA, AeVgB, AeVgC (Aedes); CpVg1a, CpVg1b, CpVg2a, CpVg2b (Culex) [30] | Genus-specific duplication patterns [30] |
| Tomato Leaf Miner (Tuta absoluta) | Insect (Lepidoptera) | 1 (TaVg characterized) | TaVg | Conserved lepidopteran Vg [17] |
| Nematode (Caenorhabditis elegans) | Nematode | 6 | vit-1 to vit-6 | Tandem duplications; vit-3/vit-4 most recent [4] |
The evolutionary history of vertebrate Vtg genes is particularly shaped by the "3R hypothesis," which posits three rounds of whole-genome duplication. A post-R3 lineage-specific duplication in teleosts gave rise to paralogous Vtg clusters that correlate with egg buoyancy—pelagic (floating) or benthic (bottom-dwelling) [29]. This adaptation was crucial for the oceanic radiation of teleosts, as the differential proteolysis of specific Vtg paralogs generates free amino acid pools that drive oocyte hydration [29].
In invertebrates, different evolutionary patterns are observed. In mosquitoes, Vtg genes evolved through duplication, concerted evolution, and purifying selection, with patterns varying by genera [30]. Culex and Aedes show evidence of gene conversion, while Anopheles Vtg genes are organized as tandem repeats, likely maintained by unequal crossover [30].
Functional Diversification of Vtg Genes
The duplication of Vtg genes has enabled functional diversification, with paralogs often acquiring specialized roles that extend beyond nutrition.
Table 2: Non-Nutritional Functions of Vitellogenins
| Organism | Documented Non-Nutritional Functions | Experimental Evidence |
|---|---|---|
| Honeybee | Behavioral regulation (foraging onset & bias), longevity, antioxidant activity, immune function [15] [4] | RNAi knockdown caused earlier foraging, nectar preference, reduced lifespan [15] |
| Mosquitoes | Potential immune, antioxidant, and metal ion transport functions [30] | Comparative genomics and sequence analysis [30] |
| Nematode (C. elegans) | Support for post-embryonic development and fertility, especially in harsh environments; intergenerational signaling [4] | Analysis of yolk composition and maternal-age effects [4] |
Case Study: Social Behavior Regulation in Honeybees
In honeybees, a single Vtg gene has been co-opted for complex social regulation. Vtg influences the age at onset of foraging, foraging specialization (nectar vs. pollen bias), and worker longevity [15]. This represents a remarkable neofunctionalization where a reproductive protein regulates behavioral castes in a eusocial insect.
The molecular mechanism involves a double repressor network with juvenile hormone (JH) [15]. Vg and JH engage in mutually inhibitory feedback, creating a regulatory circuit that paces behavioral maturation.
Figure 1: Vitellogenin Regulatory Network in Honeybees. Vg and juvenile hormone form a mutually inhibitory circuit that influences social behavior and lifespan.
RNAi-mediated knockdown of Vg confirmed these causal relationships: knockdown bees foraged earlier, showed stronger preference for nectar over pollen, and had significantly reduced lifespans [15].
Case Study: Vtg Receptors in Fish Reproduction
In flathead mullet, two distinct Vtg receptors belonging to the low-density lipoprotein receptor (LDLR) family have been identified: Lr8/VLDLR and Lrp13/LRX+1 [28]. These receptors mediate the uptake of Vtg into developing oocytes, ensuring proper yolk formation. Their characterization involved a comprehensive approach including LDLR orthology inference, protein domain analysis, 3D structure prediction, synteny evaluation, and phylogenetic analyses [28].
Experimental Approaches for Functional Validation
RNA Interference (RNAi) Protocols
RNAi has emerged as a powerful tool for validating Vtg gene function in fertility research, as demonstrated in multiple organisms.
Table 3: RNAi Experimental Protocols for Vtg Gene Functional Analysis
| Experimental Step | Honeybee (Apis mellifera) [15] | Tomato Leaf Miner (Tuta absoluta) [17] |
|---|---|---|
| dsRNA Preparation | Vg-specific dsRNA synthesized from gene sequence; GFP dsRNA as control [15] | TaVg-specific dsRNA designed from cloned gene sequence [17] |
| Delivery Method | Intra-abdominal injection into adult workers [15] | Injection into pupae [17] |
| Dosage | Not specified in abstract | Not specified in abstract |
| Validation | Vitellogenin protein level quantification (μg/μL) via Western blot [15] | qRT-PCR for TaVg expression; vitellogenin content measurement [17] |
| Functional Assays | Onset of foraging behavior, nectar vs. pollen load size, lifespan [15] | Ovarian development scoring, oocyte counts, yolk deposition, egg-laying, hatching rate [17] |
Figure 2: RNAi Experimental Workflow for Vtg Gene Functional Analysis. Standardized protocol for validating Vtg gene functions across species.
Comparative Genomics and Phylogenetic Analysis
For evolutionary studies of Vtg gene multiplicity, comparative genomic approaches have proven highly effective:
- Orthology Inference: Identify Vtg orthologs across multiple species using sequence similarity and synteny conservation [28] [30]
- Phylogenetic Analysis: Reconstruct gene trees using maximum likelihood or Bayesian methods to elucidate duplication histories [29] [30]
- Selection Tests: Detect signatures of positive or purifying selection using dN/dS ratio tests [30]
- Gene Conversion Analysis: Identify concerted evolution through statistical tests for recombination [30]
In mosquitoes, researchers identified the ancestral Vtg gene copy through conservation of a specific genomic structure—a Vtg gene associated with a upstream TRIM37-like (T37L) gene approximately 1kb away, which was conserved across all examined genera [30].
The Scientist's Toolkit: Essential Research Reagents
Table 4: Key Research Reagents for Vtg Gene Functional Studies
| Reagent/Resource | Application | Function/Rationale | Example Use |
|---|---|---|---|
| Vtg-specific dsRNA | RNAi-mediated knockdown | Silences target Vtg gene to establish causal relationships [15] [17] | Functional validation in honeybees, tomato leaf miner [15] [17] |
| Control dsRNA (e.g., GFP) | RNAi experimental control | Controls for non-specific immune responses and injection effects [15] | Honeybee behavioral studies [15] |
| Vtg Receptor Antibodies | Protein localization and quantification | Detect receptor expression in oocyte membranes [28] | Fish Vtg receptor characterization [28] |
| EST and Genomic Libraries | Gene discovery and sequence analysis | Identify Vtg gene family members and regulatory elements [31] [30] | Mosquito Vtg gene family analysis [30] |
| Species-Specific Promoter Sequences | Regulatory studies and transgenics | Drive tissue-specific expression for functional analysis [30] | Mosquito blood meal-induced activation studies [30] |
| Midazolam-d6 | Midazolam-d6, MF:C18H13ClFN3, MW:331.8 g/mol | Chemical Reagent | Bench Chemicals |
| Phytol-d5 | Phytol-d5, MF:C20H40O, MW:301.6 g/mol | Chemical Reagent | Bench Chemicals |
The multiplicity of Vtg genes and their functional diversification represents a compelling example of how gene duplication drives evolutionary innovation in reproduction. The comparative data presented in this guide reveals both conserved themes and species-specific adaptations, highlighting Vtg genes as key modules in the evolution of diverse reproductive strategies. For fertility researchers, the experimental approaches and reagents detailed here provide a roadmap for functional validation of Vtg genes and their receptors in both established and emerging model organisms. The continued exploration of Vtg gene multiplicity promises not only to advance fundamental understanding of reproductive biology but also to inform biotechnology applications in aquaculture, pest control, and conservation.
Experimental Approaches for Functional Analysis: From Gene Silencing to Biomarker Detection
RNA Interference (RNAi) for Targeted Vg Gene Knockdown
Vitellogenin (Vg), a yolk protein precursor, is a critical gene in fertility research across species. Its function extends beyond egg yolk provision to roles in oxidative stress resistance, hormonal regulation, and behavioral maturation. Validating its specific functions requires precise gene silencing techniques. RNA interference (RNAi) has emerged as a powerful tool for targeted Vg gene knockdown, enabling researchers to dissect its role in complex biological processes such as fertility. This guide objectively compares the performance of key RNAi strategies for Vg knockdown, supported by experimental data and detailed protocols.
Core RNAi Methodologies for Gene Knockdown
RNAi techniques function by introducing sequence-specific double-stranded RNA (dsRNA) that leads to the degradation of complementary messenger RNA (mRNA), thereby knocking down gene expression. The primary methods used in research are summarized below.
Table 1: Core RNAi Methodologies for Gene Knockdown
| Method | Mechanism | Key Features | Primary Applications |
|---|---|---|---|
| Long dsRNA Injection [32] | Introduction of long dsRNA (200-300+ bp) into the body cavity. Processed by Dicer into siRNAs. | Triggers specific gene silencing in insects; lacks nonspecific interferon response seen in mammals [32]. | Functional gene studies in insects (e.g., honey bees, Drosophila). |
| siRNA/siRNA Transfection [33] | Synthetic, short interfering RNAs (siRNAs; 21-nt duplexes) delivered into cells via transfection (lipids or electroporation). | Induces transient gene silencing (days) in mammalian cells; high specificity for target mRNA [33]. | Rapid, post-transcriptional gene knockdown in cultured mammalian cells. |
| shRNA Expression Vectors [34] | Vector-based expression of short hairpin RNAs (shRNAs) transcribed from a U6 promoter. Processed into siRNAs inside the cell. | Enables long-term, stable gene suppression; can be combined for multi-gene targeting (co-RNAi) [34]. | Long-term gene silencing and combinatorial RNAi in various cell types. |
| Virus-Induced Gene Silencing (VIGS) [35] | A modified virus delivers a fragment of the host plant's target gene, initiating RNAi against the host's own mRNA. | A high-throughput tool for in planta validation of gene function without stable transformation [35]. | Functional genomics in plants, including those recalcitrant to transformation (e.g., watermelon). |
The following diagram illustrates the core workflow and mechanisms shared across these RNAi methodologies.
Performance Comparison of RNAi Strategies for Vg Knockdown
Different RNAi strategies have been applied to study the Vg gene in model organisms, yielding varying efficiencies and phenotypic outcomes.
Table 2: Performance Comparison of RNAi Strategies in Vg Research
| Organism | RNAi Strategy | Knockdown Efficiency & Quantitative Data | Key Phenotypic Outcomes in Fertility |
|---|---|---|---|
| Honey Bee (Apis mellifera) | Abdominal injection of Vg-dsRNA [36] [32] | Extensive gene expression changes in the brain; Vg protein level reduction confirmed [36]. | Altered behavioral maturation; serves as a predictor for foraging, linked to social behavior [36] [32]. |
| Honey Bee (Apis mellifera) | Double Gene Knockdown (Vg + ultraspiracle) [32] | Effective simultaneous suppression of two genes [32]. | Enabled dissection of gene interrelationships and joint effects on gustatory perception and physiology [32]. |
| Fruit Fly (D. melanogaster) | Transgenic (GAL4/UAS) with honey bee Vg or fly CG31150 (Vg-like) [37] | Successful transgenic expression achieved [37]. | No increase in lifespan or fecundity; highlights functional divergence between species [37]. |
| Drosophila (D. melanogaster) | shRNA-driven depletion of AAGAG satellite RNA [38] | ~72% reduction in AAGAG RNA [38]. | 100% male sterility; defective histone-protamine exchange, abnormal sperm maturation [38]. |
Detailed Experimental Protocols
This protocol is effective for knocking down genes in adult honey bees and can be adapted for double gene knockdown.
Key Reagent Solutions:
- RiboMax T7 RNA Production System (Promega): Used for high-yield in vitro transcription of dsRNA.
- Gene-Specific Primers (designed with Primer3): Must include T7 promoter sequences at both ends to enable transcription of both sense and antisense strands.
- TRIzol-LS Reagent: Used for purifying synthesized dsRNA from the transcription reaction mixture.
- Nuclease-Free Water: Essential for dissolving the final dsRNA pellet to avoid degradation.
Methodology:
- dsRNA Synthesis:
- Design primers flanking a 200-500 bp region of the target Vg gene, with T7 promoter sequences added to the 5' ends.
- Amplify the template by PCR from cDNA. Use the PCR product in an in vitro transcription reaction with the RiboMax T7 system to produce dsRNA.
- dsRNA Purification:
- Denature and renature the dsRNA by heating to 85°C for 5 minutes and then cooling slowly to room temperature to ensure proper duplex formation.
- Treat with DNase I to remove the DNA template.
- Purify the dsRNA using TRIzol-LS and isopropyl alcohol precipitation, followed by a 75% ethanol wash. Resuspend the final pellet in nuclease-free water to a high concentration (e.g., 9-10 μg/μl).
- Abdominal Injection:
- Immobilize newly emerged honey bees by chilling at 4°C for 1-2 minutes.
- Mount bees on a wax-filled Petri dish using insect pins.
- Using a microsyringe fitted with a disposable 30G needle, inject 3 μl of purified dsRNA into the abdominal cavity. For double gene knockdown, either mix dsRNAs for a single injection or inject them on consecutive days.
This method is standard for achieving stable and specific gene knockdown in a cell culture environment.
Key Reagent Solutions:
- Synthetic siRNA Duplexes or shRNA Plasmids: 21-nucleotide RNAs for transient transfection, or vector-based shRNAs for long-term suppression.
- Lipid-Based Transfection Reagent: Facilitates the encapsulation and delivery of RNA or DNA into cells.
- Electroporation System: Applies an electrical field to create temporary pores in cell membranes for nucleic acid entry.
Methodology:
- Design: Select a 21-nt sequence from the target Vg mRNA. For shRNAs, design oligonucleotides that form a short hairpin structure with a 19-21 bp stem and a loop, often using the miR-30 loop sequence.
- Delivery:
- Transfection: Complex synthetic siRNAs or shRNA plasmids with a lipid-based transfection reagent and add to cells.
- Electroporation: Resuspend cells in a buffer containing the siRNA or shRNA plasmid and electroporate using an optimized electrical pulse.
- Analysis: Assess knockdown efficiency 48-72 hours post-delivery via qRT-PCR or Western blotting.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagent Solutions for RNAi Experiments
| Reagent / Tool | Function & Role in RNAi | Example Use Case |
|---|---|---|
| dsRNA Synthesis Kit | High-yield production of long double-stranded RNA for triggering RNAi in non-mammalian systems. | RiboMax T7 System used for generating Vg-dsRNA in honey bee studies [32]. |
| siRNA/shRNA Vectors | Plasmid-based delivery of short hairpin RNAs for stable, long-term gene silencing in cells. | pchU6-3-ClaI vector for shRNA expression; pRFPRNAiC for miRNA-embedded shRNAs [34]. |
| VIGS Vector | Modified viral vector for delivering host gene fragments into plants to induce silencing. | pCF93 vector from cucumber fruit mottle mosaic virus for high-throughput gene validation in watermelon [35]. |
| Transfection Reagent | Chemical carrier that complexes with nucleic acids to facilitate their uptake by cells. | Lipid-based reagents used for delivering siRNAs into cultured mammalian cells [33]. |
| CARD Web Tool | An integrated platform for comprehensive analysis of RNAi-screen data, including normalization and off-target analysis. | Used to increase validation rates of primary screen hits and improve concordance between screens [39]. |
| Triclocarban-13C6 | Triclocarban-13C6, MF:C13H9Cl3N2O, MW:321.5 g/mol | Chemical Reagent |
| 3,5-Dichlorobenzoic-d3 Acid | 3,5-Dichlorobenzoic-d3 Acid | 3,5-Dichlorobenzoic-d3 Acid (C7HCl2D3O2) is a stable isotope-labeled internal standard for research. This product is for Research Use Only (RUO). Not for human or veterinary use. |
Analysis of Signaling Pathways and Experimental Workflows
Knockdown of Vg can have cascading effects on interconnected signaling pathways. The diagram below synthesizes a generalized pathway based on findings from honey bee and Drosophila research, showing key molecular relationships and phenotypic outcomes.
Advanced Strategies: Combinatorial RNAi and Data Analysis
For complex biological questions, such as dissecting gene networks, advanced RNAi strategies are required.
Combinatorial RNAi (co-RNAi): This approach simultaneously knocks down multiple genes. A direct comparison of co-RNAi strategies found that multiple U6/shRNA cassettes in a single construct provided the most reliable and predictable suppression of both single and multiple-gene targets. While long hairpin RNAs (lhRNAs) and microRNA-embedded shRNAs also worked, their efficiency was inconsistent and highly dependent on siRNA position and local sequence properties [34].
Computational Analysis of Screen Data: Tools like the Comprehensive Analysis of RNAi-screen Data (CARD) platform are critical for robust data interpretation. CARD performs sequential analysis steps, including data normalization, off-target effect analysis (e.g., Common Seed Analysis), and integration with gene expression data. This significantly increases the validation rate of primary screen hits and the overlap between independent screens [39].
The choice of RNAi strategy for Vg knockdown is dictated by the experimental organism, the required duration of silencing, and the biological question. Long dsRNA injection remains the gold standard for functional studies in insects, reliably producing phenotypic effects on fertility and behavior. In mammalian systems, siRNA/shRNA technologies offer precision and flexibility. For dissecting complex genetic interactions, combinatorial RNAi using multiple shRNA cassettes is the most effective strategy. Across all systems, rigorous experimental design, including appropriate controls and computational validation of screen data, is paramount for accurately defining the role of Vg in fertility and other biological processes.
Quantitative PCR Protocols for Vtg Expression Profiling
Vitellogenin (Vtg) expression profiling serves as a critical biomarker in fertility research, providing insights into reproductive endocrine function and ovarian development. Quantitative PCR (qPCR) has emerged as the benchmark technology for precise quantification of Vtg mRNA transcripts due to its exceptional sensitivity, specificity, and wide dynamic range [40]. This technical guide provides a comprehensive comparison of RT-qPCR methodologies for Vtg expression analysis, framed within the context of validating vitellogenin gene functions in fertility studies. The selection of appropriate qPCR protocols directly impacts data reliability in investigating endocrine disruptions, assessing ovarian reserve, and understanding molecular mechanisms underlying reproductive pathologies [41]. As the field moves toward increasingly personalized approaches in fertility treatments [42], standardized and validated molecular protocols become paramount for generating comparable data across research institutions and clinical settings.
Fundamental RT-qPCR Methodologies: One-Step vs. Two-Step Approaches
The quantification of mRNA using RT-PCR can be achieved through two primary methodological frameworks: one-step and two-step amplification. The fundamental distinction lies in whether reverse transcription and PCR amplification are performed in a single combined reaction or as separate sequential reactions [43] [40].
Table 1: Comparison of One-Step and Two-Step RT-qPCR Approaches
| Parameter | One-Step RT-qPCR | Two-Step RT-qPCR |
|---|---|---|
| Workflow | cDNA synthesis and qPCR in single tube | Separate RT and qPCR reactions |
| Primer Options | Gene-specific primers only | Random hexamers, oligo-dT, or gene-specific primers |
| Hands-on Time | Minimal | Extended |
| Risk of Contamination | Low (closed-tube) | Higher (multiple open-tube steps) |
| Sample Throughput | Ideal for high-throughput | Better for multiple targets from few samples |
| RNA Input Flexibility | Limited | Flexible (can scale up RT reaction) |
| cDNA Archive Potential | No (product not saved) | Yes (cDNA stored for future assays) |
| Optimization Flexibility | Limited | High (independent optimization of RT and PCR) |
| Ideal Application | Quantitating same gene(s) across many samples | Assessing multiple targets from limited RNA samples |
One-Step RT-qPCR Protocol
The one-step protocol consolidates reverse transcription and PCR amplification into a single reaction vessel using a common buffer system. This approach employs gene-specific primers for both cDNA synthesis and amplification [43]. The methodology is particularly suitable for Vtg profiling when analyzing large sample sets targeting a limited number of genes, as it minimizes handling time and reduces cross-contamination risks [43]. Commercial one-step kits such as the Luna Universal One-Step RT-qPCR Kit (NEB #E3005) provide optimized reagent formulations for this approach [43].
Detailed Protocol:
- Reaction Setup: Combine 100-500 ng total RNA, one-step reaction buffer, reverse transcriptase, thermostable DNA polymerase, gene-specific Vtg primers (200-400 nM each), dNTPs, and optional reference dye in a single tube.
- Thermal Cycling: Program thermal cycler with reverse transcription (50-55°C for 10-30 minutes), initial denaturation (95°C for 2-10 minutes), followed by 40-45 cycles of denaturation (95°C for 10-15 seconds) and combined annealing/extension (60°C for 30-60 seconds).
- Data Collection: Acquire fluorescence during each cycle's extension phase.
Two-Step RT-qPCR Protocol
The two-step approach physically separates cDNA synthesis from PCR amplification, offering greater experimental flexibility. The initial reverse transcription can be primed using random hexamers, oligo-dT primers, gene-specific primers, or combinations thereof [43]. This method generates cDNA archives that support multiple assays from a single RNA sample, making it ideal for comprehensive Vtg expression studies alongside other fertility biomarkers.
Detailed Protocol: Step 1 - cDNA Synthesis:
- Primer Annealing: Combine 500 ng - 1 μg total RNA with RT primers (50 ng random hexamers, 25 pmol oligo-dT, or 10 pmol gene-specific primers) and nuclease-free water to 12 μL.
- Denaturation: Incubate at 65°C for 5 minutes, then immediately chill on ice.
- Reverse Transcription: Add 4 μL 5X reaction buffer, 2 μL dNTPs (10 mM), 1 μL reverse transcriptase (e.g., LunaScript RT), and 1 μL RNase inhibitor. Incubate at 25°C for 10 minutes (primer annealing), 42-55°C for 30-60 minutes (cDNA synthesis), then 70°C for 15 minutes (enzyme inactivation).
Step 2 - qPCR Amplification:
- Reaction Setup: Combine 2-5 μL diluted cDNA (1:5-1:10) with 10-12.5 μL 2X master mix (e.g., Luna Universal qPCR Master Mix), Vtg-specific primers (200 nM each), and nuclease-free water to 20-25 μL.
- Thermal Cycling: Program initial denaturation (95°C for 2-10 minutes) followed by 40-45 cycles of denaturation (95°C for 10-15 seconds) and annealing/extension (60°C for 30-60 seconds).
- Data Collection: Acquire fluorescence during each cycle's extension phase.
Critical Experimental Considerations for Vtg Quantification
RNA Quality and Normalization Strategies
RNA integrity profoundly impacts Vtg quantification accuracy. Studies comparing gene expression between fresh frozen and formalin-fixed, paraffin-embedded (FFPE) tissues demonstrate that proper normalization can compensate for degradation effects, though moderately to highly expressed genes with significant inter-sample variation correlate best [44]. For Vtg profiling in fertility research, where sample availability may be limited (e.g., ovarian biopsies), implementing rigorous RNA quality assessment using RNA Integrity Number (RIN) >9.0 is recommended [45].
Reference gene selection requires empirical validation in each experimental system. As demonstrated in drought-stress poplar studies, evaluation approaches including standard deviation analysis of Cq values, regression analysis of Cq patterns, and algorithms like NormFinder identify optimal reference genes [46]. For Vtg studies in fertility models, multiple reference genes (e.g., ACT, EF1, UBQ) should be validated to account for potential variation under experimental conditions [46].
Quantification Approaches and Data Analysis
Relative quantification of Vtg expression typically employs the comparative Cq method (2-ΔΔCq), which assumes optimal and equal amplification efficiencies for target and reference genes [46]. When amplification efficiencies differ, efficiency-corrected models like the Pfaffl method provide more accurate quantification [46].
Table 2: Comparison of Relative Quantification Methods
| Method | Principle | Efficiency Consideration | Applications |
|---|---|---|---|
| Comparative Cq (2-ΔΔCq) | Relative expression calculated from Cq differences | Assumes efficiency = 2 | High-throughput screening with validated primers |
| Pfaffl Model | Efficiency-corrected relative quantification | Uses experimentally determined efficiencies | When target/reference efficiencies differ |
| qBase Software | Multi-reference gene normalization | Incorporates efficiency parameters | High-accuracy studies requiring robust normalization |
| Standard Curve Method | Absolute quantification against serial dilutions | Direct efficiency calculation | When copy number determination required |
Amplification efficiency determination remains crucial for accurate Vtg quantification. While serial dilution methods calculate efficiency from the relationship between Cq and template concentration (E = 10(-1/slope)), this approach may overestimate efficiency due to inhibitor dilution [46]. Fluorescence curve analysis methods (e.g., LinRegPCR) determine individual reaction efficiencies from the exponential phase, potentially providing more reliable results [46].
Research Reagent Solutions for Vtg Profiling
Table 3: Essential Reagents for Vtg RT-qPCR Profiling
| Reagent Category | Specific Examples | Function in Vtg Profiling |
|---|---|---|
| One-Step Kits | Luna Universal One-Step RT-qPCR Kit (NEB #E3005) | Combined reverse transcription and amplification in single tube |
| Two-Step RT Kits | LunaScript RT SuperMix Kit (NEB #E3010) | High-efficiency cDNA synthesis with choice of priming strategies |
| qPCR Master Mixes | Luna Universal qPCR Master Mix (NEB #M3003) | Sensitive detection with intercalating dyes for Vtg amplification |
| Detection Chemistries | SYBR Green, TaqMan Probes | Fluorescence-based product detection (SYBR=economical, TaqMan=specific) |
| RNA Isolation Kits | TRIzol-based methods | High-quality RNA isolation critical for accurate Vtg quantification |
| Quality Assessment | Agilent Bioanalyzer, spectrophotometry | RNA integrity verification (RIN >9.0 recommended) |
Experimental Workflows and Pathway Diagrams
Two-Step RT-qPCR Workflow for Vtg Profiling
(Two-Step RT-qPCR Workflow for Vtg Profiling)
One-Step RT-qPCR Workflow for Vtg Profiling
(One-Step RT-qPCR Workflow for Vtg Profiling)
Selection of appropriate qPCR methodologies for Vtg expression profiling requires careful consideration of experimental objectives, sample availability, and throughput requirements. The one-step approach offers streamlined processing for high-throughput analysis of limited targets, while the two-step method provides superior flexibility for comprehensive biomarker profiling across multiple fertility endpoints. As fertility research increasingly incorporates artificial intelligence and personalized medicine approaches [42], standardized Vtg quantification protocols will play an essential role in generating reproducible data sets. Implementation of rigorous quality control measures, including RNA integrity verification, reference gene validation, and efficiency-corrected quantification, ensures reliable Vtg expression data for validating vitellogenin gene functions in reproductive biology and toxicology studies.
Standardization of Vitellogenin as an Endocrine Disruption Biomarker
The validation of vitellogenin (VTG) as a standardized biomarker represents a critical advancement in environmental endocrinology and fertility research. VTG, a yolk precursor protein normally synthesized by female oviparous vertebrates, is induced in males and juveniles upon exposure to estrogenic compounds, providing a sensitive indicator of endocrine disruption [47]. Its induction is directly linked to impaired reproductive outcomes, including skewed sex ratios, reduced fertility, and compromised hatching success, thereby bridging molecular exposure signals to adverse effects on fertility [48]. This guide objectively compares the performance of VTG as a biomarker against other alternatives, detailing experimental protocols and data underpinning its standardization within a framework focused on validating gene functions in reproductive health.
Evidence for Standardization: Performance Comparison with Alternative Biomarkers
The standardization of VTG is supported by extensive data comparing its sensitivity, specificity, and predictive value against other physiological, molecular, and histological endpoints.
Table 1: Comparative Performance of VTG and Alternative Biomarkers for Detecting Estrogenic Activity
| Biomarker | Organism | Experimental Context | Key Performance Findings | Inference for Fertility |
|---|---|---|---|---|
| Vitellogenin (VTG) | Medaka (Oryzias latipes) | 2-8 wk exposure to o,p'-DDT (0.5-7.5 ppb) [48] | Induced after 8 wk at all doses; more sensitive than gonad histology after 2 wk | Reduced fertility/hatching at all doses, even without VTG induction after 2 wk |
| Vitellogenin (vtgAb) | Spotted Scat (Scatophagus argus) | In vivo/in vitro exposure to 17α-ethynylestradiol (EE2) [49] | vtgAb most responsive subtype (30-fold in vitro); dose/time-dependent response | Direct indicator of yolk formation, crucial for oocyte maturation |
| Gonadal Histopathology | Medaka (Oryzias latipes) | 2-8 wk exposure to o,p'-DDT (0.5-7.5 ppb) [48] | Feminization/ovotestes at higher doses; progressive over time | Direct observation of impaired gonad development |
| Sex Ratio | Medaka (Oryzias latipes) | 2-8 wk exposure to o,p'-DDT (0.5-7.5 ppb) [48] | Female-skewed sex ratio at highest doses (2.5, 7.5 ppb) | Direct link to population-level reproductive failure |
| DMRT93B & Cuticle 12 | Water Flea (Daphnia magna) | Exposure to fenoxycarb (1-1000 ng/L) [50] | Significant dose-response; notable changes at 1 ng/L | Linked to developmental abnormality and reduced reproduction |
The data demonstrates that VTG induction is a highly sensitive early-warning signal. However, the case of medaka exposed to o,p'-DDT for two weeks shows that reproductive impairment (reduced fertility and hatching success) can occur even in the absence of measurable VTG induction [48]. This evidence is crucial for standardization, indicating that while VTG is a valuable biomarker, its absence does not guarantee the absence of endocrine-disrupting effects, advocating for its use within a weight-of-evidence framework alongside other endpoints like histopathology and direct fertility metrics [48] [9].
Experimental Protocols for VTG Analysis
Standardized methodologies are foundational to the reliable measurement of VTG for both research and regulatory purposes.
In Vivo Induction and Sampling in Fish Models
The protocol for medaka is representative of standardized fish bioassays [48].
- Exposure Regimen: Juvenile or male fish are exposed to the test chemical (e.g., o,p'-DDT at 0.5-7.5 ppb) in a flow-through system for a defined period (e.g., 2-8 weeks).
- Sampling: Blood is drawn from the fish, and plasma is separated via centrifugation.
- Tissue Collection: Liver tissue (the primary site of VTG synthesis) may be collected for gene expression analysis (qPCR/RNA-Seq), and gonads are preserved for histological examination to correlate VTG induction with morphological changes.
In Vitro Induction in Hepatocyte Cultures
Isolated hepatocytes provide a controlled system for mechanistic studies [49].
- Hepatocyte Isolation: Liver is aseptically dissected from the organism (e.g., S. argus), and hepatocytes are isolated via enzymatic perfusion with collagenase.
- Culture and Exposure: Cells are cultured in hormone-free medium and exposed to a range of chemical concentrations (e.g., EE2 at 10â»â· mol/L). A positive control (e.g., 17β-estradiol) and solvent control are included.
- Endpoint Analysis: After a set period (e.g., 72 hours), cells are harvested. VTG mRNA expression is quantified using qPCR, and VTG protein in the culture medium is measured by ELISA or Western blot.
VTG Quantification: ELISA and Molecular Assays
Immunoassays are the gold standard for VTG protein detection [47].
- Antibody Production: VTG is purified from the plasma of estrogen-exposed males. Polyclonal or monoclonal antibodies are then generated in a host species (e.g., rabbits).
- Sandwich ELISA:
- A capture antibody (specific to VTG) is coated onto a microtiter plate.
- Plasma samples or standards are added. VTG binds to the capture antibody.
- A detection antibody (also VTG-specific, often conjugated to an enzyme like horseradish peroxidase) is added.
- A substrate solution is added, and the enzymatic reaction produces a color change measurable with a spectrophotometer. The intensity is proportional to the VTG concentration.
For gene expression studies, RNA is extracted from liver tissue or hepatocytes, reverse-transcribed to cDNA, and analyzed via qPCR using primers specific to VTG genes or subtypes (e.g., vtgAa, vtgAb, vtgC) [49].
Molecular Mechanisms and Standardized Workflow
The VTG induction pathway is a canonical estrogen receptor-mediated process. Standardizing its use requires understanding this mechanism and implementing a consistent experimental workflow.
Diagram 1: Molecular Signaling Pathway of VTG Induction. EDCs activate estrogen receptors, triggering transcription of the vtg gene and leading to VTG protein synthesis, a biomarker linked to fertility impairment.
The visualization above depicts the core molecular pathway. Estrogenic EDCs, such as EE2 or nonylphenol, enter the hepatocyte and bind to nuclear estrogen receptors (ERα/ERβ) [9]. This ligand-receptor complex then binds to estrogen response elements (EREs) on the DNA, activating the transcription of vtg genes. The resulting VTG mRNA is translated into VTG protein, which is secreted into the bloodstream. In males, this protein serves as a definitive biomarker of exposure. The integration of this mechanism with other omics data (transcriptomic, epigenetic) places VTG within a broader Adverse Outcome Pathway (AOP) framework, linking molecular initiation to population-level effects [9].
Diagram 2: Standardized Experimental Workflow for VTG Biomarker Application. The process from model selection to data integration ensures consistent and reproducible assessment of endocrine disruption.
The Scientist's Toolkit: Essential Research Reagents and Materials
The following table details key reagents and their functions for conducting VTG-based endocrine disruption research.
Table 2: Essential Research Reagents for VTG-Based Endocrine Disruption Studies
| Reagent / Material | Function in Research | Application Example |
|---|---|---|
| 17α-Ethynylestradiol (EE2) | Potent synthetic estrogen; positive control for exposure studies [9] [49] | Inducing VTG in male fish hepatocyte cultures [49] |
| Species-Specific VTG Antibodies | Core component of immunoassays (ELISA, Western Blot) for VTG protein detection and quantification [51] [47] | Measuring VTG levels in plasma of male fish exposed to effluent [51] |
| qPCR Primers for vtg Subtypes | Enable quantification of vtg gene expression levels (mRNA) and discrimination between subtypes [49] | Identifying vtgAb as the most EE2-responsive subtype in S. argus [49] |
| Primary Hepatocyte Culture System | In vitro model for mechanistic studies, reducing whole-animal use [49] | Testing direct estrogenicity of chemicals on liver cells [49] |
| RNAi Reagents (dsRNA) | Gene silencing tool for functional validation of vtg and its receptor (vtgR) in fertility [17] | Silencing TaVg in T. absoluta to confirm its role in oocyte development and fecundity [17] |
| Desglycolaldehyde Desonide | Desglycolaldehyde Desonide, MF:C22H30O4, MW:358.5 g/mol | Chemical Reagent |
| 3-Hydroxynortriptyline | 3-Hydroxynortriptyline | 3-Hydroxynortriptyline is a research metabolite of the antidepressant Nortriptyline. This product is For Research Use Only. Not for human or veterinary use. |
The standardization of vitellogenin as a biomarker for endocrine disruption is robustly supported by its sensitive and quantifiable induction in male and juvenile fish, its conserved mechanistic pathway, and its clear link to adverse reproductive outcomes. Critical insights, such as the dissociation between VTG induction and reproductive impairment in some scenarios, underscore the necessity of employing VTG within a integrated biomarker strategy. Future directions involve refining sex-specific biomarker panels, incorporating multi-omics data, and leveraging machine learning to enhance predictive risk assessment, ultimately strengthening the validation of vitellogenin gene functions in fertility research and environmental safety.
Multi-omics integration represents a transformative approach in molecular biology, enabling researchers to simultaneously profile multiple molecular layers from the same biological sample. This powerful strategy combines transcriptomic, epigenomic, and proteomic data to construct comprehensive molecular profiles that link gene regulation, transcriptional output, and protein function [52]. In the specific context of vitellogenin (Vg) gene validation in fertility research, multi-omics approaches provide unprecedented opportunities to decipher the complex regulatory mechanisms governing reproductive biology. Vitellogenin, a glycolipid complex protein produced in the fat body of female insects, plays vital roles in oocyte development and reproductive regulation [17]. The integration of transcriptomic, epigenetic, and proteomic analyses creates a powerful framework for validating Vg gene functions and understanding its role in fertility across species.
The power of multi-modal integration is particularly valuable in fertility research, where it enables researchers to connect epigenetic regulatory mechanisms with transcriptional outputs and functional protein consequences. For example, in studying Vg-mediated reproduction, combining chromatin accessibility data with gene expression profiles can reveal regulatory elements driving Vg transcription, while protein quantification confirms the translational products critical for ovarian development [52]. This integrated perspective is essential for comprehensive validation of gene functions in complex biological processes like reproduction, where multiple regulatory layers interact to determine phenotypic outcomes.
Comparative Analysis of Multi-omics Technologies and Platforms
Technology Platform Comparison
Table 1: Comparison of major single-cell multi-omics platforms
| Platform/Company | Key Technology | Omics Layers Captured | Best Application Scenarios | Throughput Considerations |
|---|---|---|---|---|
| 10x Genomics Multiome | Single-cell RNA + ATAC sequencing | Transcriptome + Epigenome (chromatin accessibility) | High-throughput genomics-focused studies, large sample sizes | High scalability for large studies [53] |
| Mission Bio Tapestri | Targeted DNA sequencing + protein detection | Genome + Proteome | Targeted mutation studies, tumor heterogeneity, rare cell detection | Ideal for focused panels [53] |
| BD Rhapsody | Whole transcriptome + protein tagging | Transcriptome + Proteome | Immunology research, flexible workflow needs | Moderate to high throughput [53] |
| CITE-seq | Oligonucleotide-tagged antibodies + RNAseq | Transcriptome + Proteome | Immune profiling, surface marker validation | Compatible with various seq platforms [52] |
| TEA-seq | Multimodal cell profiling | Transcriptome + Epigenome + Proteome | Comprehensive cell typing, deep phenotyping | Lower throughput but complete data [52] |
Bioinformatics Integration Tools
Table 2: Multi-omics data integration tools and their applications
| Integration Method | Representative Tools | Strengths | Limitations | Suitable for Vg Research |
|---|---|---|---|---|
| Network Propagation/Diffusion | MOFA [52] | Captures global data patterns, handles missing data | Complex interpretation, computational demands | Moderate for pathway analysis |
| Similarity-based Approaches | Harmony [52] | Efficient for large datasets, batch correction | May oversimplify biological complexity | Good for cross-species comparison |
| Graph Neural Networks | Various custom implementations | Models complex interactions, high accuracy | Requires large training datasets, "black box" | Emerging potential |
| Network Inference Models | Seurat [52] | Cell type identification, spatial mapping | Primarily for single-cell data | High for single-cell Vg studies |
Experimental Design for Vitellogenin Gene Function Validation
Integrated Multi-omics Workflow for Vitellogenin Studies
The validation of vitellogenin gene functions in fertility research requires a carefully designed multi-omics workflow that connects molecular measurements across different regulatory layers. Below is a standardized experimental protocol for comprehensive Vg functional analysis:
Detailed Methodologies for Vitellogenin Multi-omics Analysis
Epigenetic Profiling of Vitellogenin Regulatory Regions
Experimental Protocol:
- Sample Preparation: Isolate reproductive tissues (ovaries, fat body) from female organisms at different reproductive stages. For Tuta absoluta studies, ovarian developmental staging should be standardized prior to analysis [17].
- Chromatin Accessibility Mapping: Perform ATAC-seq (Assay for Transposase-Accessible Chromatin using sequencing) using 50,000 cells per sample. Use frozen tissues digested with collagenase to prepare single-cell suspensions. Transpose with Tn5 transposase for 30 minutes at 37°C, then purify DNA for library preparation [52].
- Histone Modification Analysis: Conduct ChIP-seq (Chromatin Immunoprecipitation followed by sequencing) for H3K27ac (activation mark) and H3K27me3 (repression mark) in Vg promoter regions. Cross-link cells with 1% formaldehyde, sonicate chromatin to 200-500 bp fragments, immunoprecipitate with specific antibodies, and sequence enriched DNA [54].
- DNA Methylation Analysis: Utilize whole-genome bisulfite sequencing or reduced representation bisulfite sequencing to profile CpG methylation in Vg gene regulatory regions. Treat DNA with sodium bisulfite, which converts unmethylated cytosine to uracil while leaving methylated cytosine unchanged [55].
Quality Control Parameters:
- Sequence depth: ≥20 million reads per sample for ATAC-seq
- Antibody validation: Verify ChIP-seq antibody specificity using knockout controls
- Bisulfite conversion efficiency: >99% for methylation studies
- Include spike-in controls for normalization
Transcriptomic Analysis of Vitellogenin Expression
Experimental Protocol:
- RNA Extraction and Quality Control: Isolve total RNA from reproductive tissues using TRIzol reagent with DNase I treatment. Assess RNA integrity using Bioanalyzer (RIN ≥8.0 required) [56].
- Library Preparation and Sequencing: Use poly-A selection for mRNA enrichment or ribosomal RNA depletion. Prepare libraries using Illumina Stranded mRNA Prep kit. Sequence on Illumina platforms with ≥30 million paired-end reads (2×150 bp) per sample [56].
- Single-Cell RNA Sequencing: For cellular resolution, prepare single-cell suspensions from reproductive tissues. Use 10x Genomics Chromium platform for cell partitioning and barcoding. Target 5,000-10,000 cells per sample with ≥50,000 reads per cell [52].
- Spatial Transcriptomics: For tissue context, utilize Visium Spatial Gene Expression platform to map Vg expression within ovarian architecture. Cryosection tissues at 10μm thickness onto capture areas [54].
Data Analysis Pipeline:
- Alignment: STAR aligner to reference genome
- Quantification: FeatureCounts or HTSeq for gene-level counts
- Differential expression: DESeq2 or edgeR with FDR <0.05
- Single-cell analysis: Seurat or Scanpy for clustering and marker identification
Proteomic Validation of Vitellogenin Protein
Experimental Protocol:
- Protein Extraction and Digestion: Homogenize tissues in RIPA buffer with protease inhibitors. Quantify protein using BCA assay. Digest 100μg protein with trypsin (1:50 ratio) overnight at 37°C [57].
- Mass Spectrometry Analysis: Desalt peptides and analyze by LC-MS/MS on Orbitrap platforms. Use data-independent acquisition (DIA) for quantitative profiling. Include retention time standardization using iRT kits [57].
- Immunoassay Validation: Implement Nu.Q immunoassays or ELISA for specific Vg quantification. Use validated antibodies against conserved Vg domains. Include standard curves with recombinant Vg protein for quantification [57].
- CITE-seq Integration: For single-cell protein detection, use oligonucleotide-tagged antibodies against Vg (10-20 antibodies per panel). Incubate with single-cell suspensions before partitioning [52].
Quantification Standards:
- MS1 normalization: Use total peptide amount or spike-in standards
- Immunoassay precision: Intra-assay CV <10%, inter-assay CV <15%
- Limit of detection: <1 ng/mL for immunoassays
The Scientist's Toolkit: Essential Research Reagents and Platforms
Table 3: Key research reagent solutions for multi-omics fertility research
| Category | Specific Product/Platform | Application in Vg Research | Technical Considerations |
|---|---|---|---|
| Epigenetic Analysis | Illumina NovaSeq 6000 | ATAC-seq for chromatin accessibility | High throughput, requires nuclear isolation |
| Nu.Q Immunoassays [57] | Histone PTM quantification | Validated for specific modifications, high throughput | |
| CUT&Tag kits | Histone mark profiling | Lower input requirements than ChIP-seq | |
| Transcriptomic Analysis | 10x Genomics Single Cell RNA-seq | Cellular resolution of Vg expression | Cell viability critical (>90%) |
| SMART-seq HT kits | Full-length RNA sequencing | Higher sensitivity for low-abundance transcripts | |
| TempO-Seq | Targeted transcriptomics | Cost-effective for large screens | |
| Proteomic Analysis | Olink Target panels | Multiplex protein quantification | Pre-validated assays, low sample volume |
| CITE-seq antibodies [52] | Surface protein + RNA sequencing | Antibody validation essential | |
| TMTpro 16-plex | Multiplex mass spectrometry | Enables 16-sample simultaneous processing | |
| Functional Validation | dsRNA synthesis kits | RNAi-mediated Vg silencing [17] | Target-specific sequence design critical |
| CRISPR-Cas9 systems | Gene editing for functional validation | Guide RNA design for minimal off-target effects | |
| 3-Phenylhexanoic acid | 3-Phenylhexanoic Acid|RUO | 3-Phenylhexanoic acid is a phenyl-substituted carboxylic acid for research use. This compound is For Research Use Only. Not for human use. | Bench Chemicals |
| 3-Ethynylpiperidin-3-ol | 3-Ethynylpiperidin-3-ol, MF:C7H11NO, MW:125.17 g/mol | Chemical Reagent | Bench Chemicals |
Case Study: Multi-omics Validation of Vitellogenin in Tuta absoluta Reproduction
Experimental Design and Multi-omics Integration
A comprehensive study on Tuta absoluta demonstrated the power of multi-omics approaches for validating Vg gene functions in reproduction [17]. The research employed an integrated methodology to connect transcriptional regulation, protein function, and phenotypic outcomes:
Quantitative Outcomes of Vitellogenin Perturbation
Table 4: Multi-omics measurements of vitellogenin functional validation
| Experimental Parameter | Control Group | dsTaVg-Treated | Experimental Method | Biological Impact |
|---|---|---|---|---|
| TaVg Transcript Level | Normalized to 1.0 | 0.32 ± 0.05 (68% reduction) | qRT-PCR | Significant gene silencing achieved [17] |
| Vitellogenin Protein | 100% ± 5% | 45% ± 8% reduction | Immunoassay quantification | Reduced yolk deposition [17] |
| Ovarian Tube Length | 1.52 ± 0.11 cm | 0.93 ± 0.09 cm | Morphometric analysis | Impaired ovarian development |
| Oocytes per Female | 185 ± 15 | 72 ± 12 | Microscopic counting | 61% reduction in egg production |
| Egg Hatching Rate | 89% ± 3% | 42% ± 7% | Fertility tracking | Severe reproductive impairment |
| Co-silencing (TaVg + VgR) | Normal fertility | 15% ± 5% egg hatching | Combined RNAi approach | Synergistic effect on fertility reduction |
The integrated multi-omics data from this study demonstrated that Vg silencing directly impacts reproductive capacity through coordinated molecular and phenotypic effects. The significant reduction in both transcript and protein levels (68% and 45% respectively) directly correlated with morphological changes in ovarian development and functional impairments in fertility metrics [17]. This case study exemplifies how multi-omics approaches can establish direct causal relationships between gene function and reproductive outcomes.
Data Integration Strategies and Computational Approaches
Network-Based Integration for Biological Insight
Advanced computational methods are essential for integrating multi-omics data to extract meaningful biological insights about vitellogenin function in fertility. Network-based integration approaches have shown particular promise for connecting disparate omics layers:
Network-based multi-omics integration methods can be categorized into four primary types, each with distinct advantages for fertility research [58]:
Network Propagation/Diffusion: These methods simulate information flow through biological networks, ideal for identifying downstream effects of Vg manipulation on reproductive pathways.
Similarity-Based Approaches: These compute similarity measures between omics profiles across samples, useful for identifying conserved Vg regulatory patterns across species.
Graph Neural Networks: Deep learning approaches that model complex non-linear relationships between different omics layers, capable of predicting novel Vg interactions.
Network Inference Models: These reconstruct regulatory networks from omics data, potentially revealing novel transcription factors controlling Vg expression during reproduction.
Cross-Species Conservation Analysis of Vitellogenin Regulation
The phylogenetic conservation of vitellogenin genes across species provides opportunities for comparative multi-omics analyses. Studies have shown that Vg genes cluster into distinct clades within Lepidoptera species, suggesting evolutionary conservation of regulatory mechanisms [17]. Multi-omics approaches can leverage this conservation to identify core regulatory features:
Table 5: Cross-species vitellogenin regulatory elements identified through multi-omics
| Regulatory Element | Species Conservation | Epigenetic Signature | Functional Role in Fertility |
|---|---|---|---|
| Vg Promoter Region | Conserved in Lepidoptera | H3K4me3, H3K27ac marks | Transcriptional activation during reproduction |
| Estrogen Response Elements | Vertebrates [48] | DNA hypomethylation | Response to estrogenic compounds |
| 5' UTR Regulatory Motifs | Insects to vertebrates | Accessible chromatin (ATAC-seq) | Translational regulation and efficiency |
| 3' UTR Stability Elements | Species-specific variations | RNA-binding protein sites | mRNA stability and degradation control |
This comparative approach demonstrates how multi-omics data integration can reveal both conserved and species-specific aspects of vitellogenin regulation, providing insights for both basic reproductive biology and applied pest management strategies [17].
The integration of transcriptomic, epigenetic, and proteomic analyses provides a powerful framework for validating vitellogenin gene functions in fertility research. The comparative analysis presented in this guide demonstrates that multi-omics approaches can establish direct causal relationships between molecular manipulations and reproductive outcomes, as evidenced by the Tuta absoluta case study where Vg silencing resulted in 68% transcript reduction and 61% decrease in egg production [17].
Future developments in multi-omics technologies will further enhance vitellogenin research through improved single-cell resolution, spatial mapping capabilities, and computational integration methods. Emerging techniques such as spatial transcriptomics and proteomics will enable researchers to precisely localize Vg expression within ovarian tissues, while advances in CRISPR-based screening technologies will facilitate high-throughput functional validation of Vg regulatory elements [54]. As these technologies mature and become more accessible, multi-omics integration will continue to transform our understanding of reproductive biology and accelerate the development of interventions targeting fertility regulation across species.
High-Throughput Screening Applications in Drug Development and Ecotoxicology
High-Throughput Screening (HTS) represents a foundational methodology in modern biological sciences, enabling the rapid experimental testing of thousands to millions of chemical or biological compounds against selected targets. This approach has become a standard tool in pharmaceutical research and environmental toxicology, dramatically accelerating the pace of discovery and assessment. HTS operates on the principle of automation and miniaturization, utilizing robotics, detectors, and sophisticated software to conduct large numbers of analyses in remarkably short timeframes [59]. The technology has evolved substantially since its widespread adoption began two decades ago, with current systems capable of examining over 100,000 compounds per day through highly parallelized processes [59].
The relevance of HTS extends across multiple domains, from initial drug discovery to toxicity assessment, with its applications continually expanding into new areas of research. In the specific context of vitellogenin gene function validation in fertility research, HTS provides powerful tools for elucidating the complex roles these genes play in reproductive biology across model organisms. Vitellogenins, as the principal yolk proteins in oviparous animals, are essential for provisioning developing embryos, and their precise functions are now being unraveled through large-scale screening approaches [4]. This guide examines how HTS methodologies are being applied in both drug development and ecotoxicology, with particular attention to their utility in fertility research centered on vitellogenin gene functions.
HTS Technological Fundamentals
Core Principles and Implementation
High-Throughput Screening relies on several interconnected technological components that together enable rapid, automated testing at scale. The system begins with assay design and miniaturization, where biological tests are optimized for small volumes and high density formats. Modern HTS typically employs microplates ranging from 96 to 1586 wells per plate, with working volumes as low as 2.5-10 μL [59]. This miniaturization significantly reduces reagent consumption and costs while increasing throughput. The second critical component is automation and robotics, which handles liquid dispensing, plate manipulation, and process regulation without manual intervention [59] [60]. These robotic systems are equipped with precision grippers that transfer microplates between different processing stations on an integrated platform.
Detection technologies form the third pillar of HTS infrastructure, with fluorescence, luminescence, and mass spectrometry being among the most common readout methods [60]. Fluorescence-based detection is particularly prominent due to its sensitivity, responsiveness, and adaptability to HTS formats. Finally, sophisticated data management and analysis systems process the enormous datasets generated, employing statistical quality control, cheminformatics, and increasingly, machine learning algorithms to identify true hits while minimizing false positives [61] [60]. The integration of these components enables the distinctive capabilities of HTS—speed, scale, and reproducibility—that have made it indispensable to contemporary biological research.
Advanced HTS Modalities
Recent technological advances have expanded HTS capabilities beyond traditional approaches. Ultra-High-Throughput Screening (uHTS) can conduct millions of assays daily, utilizing even higher density microplates (up to 3456 wells) with volumes of 1-2 μL [59] [60]. This extreme miniaturization presents technical challenges in fluid handling but offers unprecedented screening capacity. Cell-based assays have emerged as the dominant HTS format, representing 39.4% of the technology category in the HTS market [62]. These assays provide more physiologically relevant data by assessing compound effects in living systems rather than isolated biochemical preparations.
Label-free technologies represent another significant advancement, eliminating the need for fluorescent or other tags that might interfere with biological function. Additionally, lab-on-a-chip approaches and high-content screening methods combine HTS with detailed phenotypic analysis through automated microscopy and image analysis [63]. These multidimensional approaches can simultaneously capture multiple parameters from each sample, generating rich datasets that reveal subtle biological effects. The compaRe toolkit exemplifies innovations addressing the analytical challenges posed by these complex datasets, providing quality control, bias correction, and similarity analysis for multiparameter screening data [61].
Table 1: Core HTS Technologies and Characteristics
| Technology | Throughput Capacity | Key Applications | Advantages | Limitations |
|---|---|---|---|---|
| Traditional HTS | 10,000-100,000 compounds/day | Primary screening, target identification | Established protocols, robust data | Lower throughput than uHTS |
| Ultra-HTS | >100,000-1,000,000 compounds/day | Large library screening, repurposing | Maximum coverage of chemical space | High infrastructure costs, complex fluid handling |
| Cell-Based Assays | Varies with readout | Toxicology, functional screening, phenotypic analysis | Physiological relevance, pathway context | More variables to control, potential variability |
| Lab-on-a-Chip | Varies with design | Specialized applications, point-of-care | Minimal reagent use, integrated processes | Custom designs often needed |
| Label-Free Technologies | Moderate to high | Binding studies, functional responses | No label interference, real-time monitoring | May require specialized instruments |
HTS in Drug Discovery and Development
Applications and Workflows
In pharmaceutical research, HTS serves as a critical engine for early drug discovery, enabling the identification of novel therapeutic candidates from vast chemical libraries. The primary application lies in primary screening, which accounts for 42.7% of HTS applications in the market [62]. This initial phase rapidly tests thousands to millions of compounds against biological targets to identify "hits" – substances showing desired activity. These hits then progress to secondary screening, where more quantitative biological assays determine potency (e.g., IC50 values) and selectivity [59]. The target identification segment represents another growing application, with a projected CAGR of 12% through 2035 [62], underscoring how HTS accelerates the mapping of compounds to their biological targets.
The drug discovery HTS workflow typically begins with library preparation, where compound collections are formatted in microplates using automated systems [60]. Assay development follows, optimizing biological reactions for miniaturized formats while maintaining robustness and reproducibility. After automated screening execution, data analysis pipelines process results, applying statistical methods and algorithms to distinguish true signals from artifacts. A significant innovation in this domain is the "fast to failure" approach, where HTS quickly eliminates unsuitable candidates rather than just identifying promising ones, potentially saving substantial resources in later development stages [60].
Experimental Protocols in Drug Discovery
A representative HTS protocol for enzyme inhibition screening illustrates the standardized yet adaptable nature of these approaches. The procedure begins with preparation of assay components: the target enzyme diluted in appropriate buffer, test compounds dissolved in DMSO, and substrate solutions. For a fluorescence-based assay, this might involve a peptide substrate coupled to a fluorescent leaving group [60]. The protocol proceeds with automated liquid handling steps: (1) transfer of 2μL compound solutions to 1536-well plates using nanoliter dispensers, (2) addition of 10μL enzyme solution followed by incubation, and (3) addition of 10μL substrate to initiate reaction.
Following reaction completion, detection and analysis occur through plate readers measuring fluorescence intensity. Data processing normalizes signals against positive controls (no inhibition) and negative controls (complete inhibition), with hits identified as compounds showing significant signal reduction. Counter-screening then eliminates compounds acting through interference mechanisms (e.g., aggregation, fluorescence quenching) rather than specific inhibition [60]. This systematic approach enables reproducible screening of vast compound libraries while controlling for common artifacts that historically plagued HTS campaigns.
HTS Drug Discovery Workflow
HTS in Ecotoxicology and Environmental Testing
Applications and Approaches
The application of HTS in ecotoxicology has transformed environmental risk assessment by enabling rapid evaluation of chemical effects across diverse species and endpoints. Regulatory agencies like the U.S. EPA now employ HTS to prioritize chemicals for more extensive evaluation, limiting animal testing while efficiently screening thousands of compounds [64]. The Tox21 program, a collaborative effort among public health agencies, exemplifies this approach, using in vitro assays to profile compounds in a high-throughput, concentration-responsive manner [60]. These methodologies provide mechanistic insights into toxicity pathways at a scale impossible with traditional whole-organism testing.
A particularly innovative application of HTS in ecotoxicology is the HiTEC (High-Throughput Ecotoxicology) initiative, which adapts human toxicology assays for environmentally relevant species [63]. This approach includes applying "Cell Painting" or high-throughput phenotypic profiling to fish and insect cells, enabling chemical screening at higher throughput than current whole-organism methods. Cell Painting uses multiplexed fluorescent dyes to capture detailed morphological information, generating rich phenotypic profiles that can reveal subtle toxic effects [63]. This methodology allows testing in taxa unavailable for in vivo studies (e.g., reptiles, marine mammals) while providing deeper mechanistic insights than traditional viability assays.
Vitellogenin Screening in Ecotoxicology
Vitellogenin has emerged as a sensitive biomarker for estrogenic exposure in aquatic organisms, with HTS methods enabling efficient monitoring of environmental contaminants. In male fish, vitellogenin production is normally negligible but can be strongly induced by estrogen-mimicking compounds, making it a valuable indicator of endocrine disruption [8]. HTS approaches for vitellogenin detection typically utilize immunoassays or transcriptomic methods formatted for microplates, allowing parallel assessment of numerous samples across concentration gradients.
The experimental protocol for vitellogenin-based endocrine disruption screening involves cell-based reporter assays incorporating estrogen response elements linked to measurable outputs (e.g., luciferase). Alternatively, ex vivo liver explant cultures from model fish species like zebrafish can be exposed to test compounds, with vitellogenin mRNA or protein secretion quantified as the endpoint [8]. These approaches provide concentration-response data that benchmark compound potency relative to natural estrogens. However, research indicates important nuances in interpretation; for example, a study on cunner fish demonstrated that while male vitellogenin induction reliably indicates exposure to estrogens, it does not consistently predict reproductive impairment in all species [8]. This highlights the importance of complementary endpoints in HTS ecotoxicology screening.
Table 2: HTS Applications in Drug Development vs. Ecotoxicology
| Parameter | Drug Development | Ecotoxicology |
|---|---|---|
| Primary Goal | Identify therapeutic candidates | Assess environmental hazard |
| Typical Assay Formats | Target-based, cell-based phenotypic | Cell-based, zebrafish embryos, in vitro biomarkers |
| Key Endpoints | Target engagement, efficacy, cytotoxicity | Acute toxicity, endocrine disruption, neurotoxicity |
| Library Composition | Diverse synthetic compounds, natural products | Environmental contaminants, pesticides, industrial chemicals |
| Concentration Range | Typically μM-nM for efficacy | Broad range including environmental relevance |
| Model Systems | Human enzymes, cell lines, sometimes animal models | Environmental species (fish, invertebrates), ecosystems |
| Regulatory Context | FDA/EMA guidelines for drug approval | EPA, OECD guidelines for chemical safety |
| Vitellogenin Application | Limited direct application | Key biomarker for endocrine disruption screening |
Vitellogenin Gene Function Validation in Fertility Research
Vitellogenin Biology and Research Models
Vitellogenins constitute a highly conserved family of yolk proteins that serve as the primary nutrient source for developing embryos in oviparous species [4]. These large lipo-glyco-phosphoproteins are synthesized in somatic tissues (typically liver in vertebrates, intestine in nematodes), secreted into circulation, and absorbed by oocytes via receptor-mediated endocytosis. In the model nematode Caenorhabditis elegans, six vitellogenin genes (vit-1 through vit-6) are among the most highly expressed in the adult hermaphrodite intestine, producing copious yolk that provisions eggs [4]. Zebrafish possess an even more complex system with eight vitellogenin genes categorized into three types, each with distinct structural characteristics and potentially different functional roles during development [65].
The abundance and regulation of vitellogenins make them ideal subjects for HTS approaches in fertility research. In C. elegans, vitellogenins are not merely passive nutrient carriers but appear to function as intergenerational signaling molecules, communicating maternal physiological status to offspring [4]. This role is evidenced by observations that vitellogenin provisioning increases with maternal age and under certain environmental conditions, with functional consequences for progeny development [4]. Similarly, in zebrafish, different vitellogenin types contribute uniquely to reproduction and embryonic development, with type I vitellogenins essential for early embryonic and late larval development, while type III vitellogenin is critical specifically for early embryogenesis [65].
HTS Approaches for Vitellogenin Function Analysis
High-Throughput Screening methodologies enable systematic functional analysis of vitellogenin genes and their roles in fertility. In zebrafish, CRISPR/Cas9-mediated mutagenesis coupled with high-throughput phenotyping has revealed essential functions for specific vitellogenin genes. For example, disruption of vtg2 (a type II vitellogenin) causes significant mortality at early embryonic stages, with only 29% survival at 24 hours post-fertilization declining to 18% by 20 days post-fertilization [65]. Mutant embryos exhibit vitelline membrane deficiencies, yolk leakage, and morphological abnormalities, indicating Vtg2's critical role in membrane integrity beyond nutritional provision.
The experimental workflow for vitellogenin HTS in fertility research typically begins with genetic perturbation (CRISPR, RNAi) followed by automated phenotyping. In a representative study targeting zebrafish vtg2 [65], researchers designed guide RNAs to introduce a large deletion mutation, then used automated microscopy to document developmental phenotypes in F3 offspring. High-throughput image analysis quantified fertilization rates, hatching success, and developmental abnormalities, while label-free LC-MS/MS proteomics identified differentially abundant proteins in mutant versus wild-type embryos [65]. This integrated approach revealed that vtg2 mutation dysregulates proteins involved in cell cycle, protein degradation, lectins, and zona pellucida proteins, providing mechanistic insights into its reproductive functions.
Vitellogenin Functions in Fertility
Comparative Analysis and Research Applications
Cross-Domain HTS Implementation
While drug development and ecotoxicology apply HTS to different primary objectives—therapeutic discovery versus environmental safety assessment—they share common technological foundations and face similar methodological challenges. Both fields increasingly prioritize cell-based assay systems that provide greater physiological relevance than isolated biochemical preparations [62]. Both must address the persistent challenge of false positives and false negatives, employing statistical triage and confirmatory assays to distinguish true signals from artifacts [60]. Additionally, both domains are experiencing rapid growth in data complexity, necessitating advanced computational tools like the compaRe platform for multidimensional data analysis [61].
Important distinctions emerge in how HTS is applied across these domains. Drug discovery typically focuses on identifying specific molecular interactions between compounds and defined targets, while ecotoxicology often assesses broader phenotypic consequences of exposure. Consequently, ecotoxicology HTS more frequently employs diverse model organisms and complex endpoints like vitellogenin induction, which integrates multiple biological pathways [8] [63]. The concentration ranges also differ substantially, with drug discovery testing primarily at pharmacologically relevant concentrations (μM-nM), while ecotoxicology spans from environmental relevance to overt toxicity.
Essential Research Tools and Reagents
The successful implementation of HTS for vitellogenin research and related fertility studies requires specialized reagents and tools that enable precise, reproducible screening. The following table summarizes key solutions that form the foundation of these research applications.
Table 3: Essential Research Reagent Solutions for Vitellogenin and Fertility HTS
| Reagent/Tool | Function | Application Examples |
|---|---|---|
| CRISPR/Cas9 Gene Editing Systems | Targeted gene disruption for functional studies | vtg2 mutation in zebrafish to assess fertility impact [65] |
| Vitellogenin Antibodies | Protein detection and quantification in immunoassays | Measuring Vtg induction in male fish for endocrine disruption screening [8] |
| Cell Painting Kits | Multiplexed morphological profiling with fluorescent dyes | HiTEC ecotoxicology screening in fish cells [63] |
| Automated Liquid Handling Systems | Precise nanoliter dispensing for miniaturized assays | Compound library screening in 1536-well formats [60] |
| High-Content Screening Microscopes | Automated image acquisition and analysis for phenotypic screening | Developmental defect assessment in zebrafish embryos [65] |
| LC-MS/MS Systems | Protein identification and quantification via mass spectrometry | Proteomic analysis of vtg2-mutant zebrafish embryos [65] |
| Reporter Assay Kits | Transcriptional activity measurement via luciferase/fluorescence | Estrogen receptor activation screening [60] |
| 3D Culture Matrices | Support for complex tissue models in screening | Liver organoid cultures for vitellogenin production studies |
High-Throughput Screening has established itself as a transformative methodology across biological research domains, with particularly significant applications in drug development and ecotoxicology. The technology enables rapid, systematic investigation of chemical-biological interactions at scales impossible through traditional approaches. In the specific context of vitellogenin gene function validation for fertility research, HTS provides powerful tools for elucidating the complex roles these genes play in reproduction and development. From CRISPR-based functional screening to automated phenotypic analysis, HTS approaches are uncovering both the nutritional and signaling functions of vitellogenins across model organisms.
The continued evolution of HTS technologies—including further miniaturization, enhanced detection modalities, and more sophisticated data analysis platforms—promises to deepen these research applications while expanding into new areas. The growing HTS market, projected to increase from USD 32.0 billion in 2025 to USD 82.9 billion by 2035 [62], reflects the expanding adoption and capability of these approaches. As vitellogenin research increasingly incorporates these tools, our understanding of fertility mechanisms and their disruption by environmental factors will grow correspondingly, potentially enabling new interventions for both reproductive medicine and environmental protection.
Overcoming Technical Challenges: Ensuring Reproducibility in Vtg Functional Studies
Minimizing Interlaboratory Variability in Vtg Quantification
Vitellogenin (Vtg), an egg yolk precursor protein, serves as a highly sensitive biomarker for assessing estrogenic exposure in aquatic organisms and has growing relevance in understanding endocrine-disrupting effects on reproductive health [66]. In fertility research, validating biomarkers like Vtg requires exceptional precision across laboratories to ensure reproducible findings. However, interlaboratory studies have demonstrated that Vtg gene (vtg) expression monitoring programs face significant reproducibility challenges, with high biological variability observed between identically treated individuals even under controlled laboratory conditions [66] [67]. This variability, with coefficients of variation exceeding 100% in some studies, complicates the interpretation of exposure effects and hinders the validation of Vtg's specific gene functions in reproductive toxicology [67]. The quantification of Vtg transcriptional responses through quantitative real-time polymerase chain reaction (QPCR) assays presents both technical and analytical challenges that must be systematically addressed to produce reliable, comparable data across research institutions. This guide objectively compares experimental approaches and identifies critical factors that minimize variability, providing researchers with evidence-based protocols for robust Vtg quantification in fertility and toxicological research contexts.
Comparative Analysis of Vtg Quantification Approaches
Table 1: Identified Sources of Variability in Vtg Transcript Quantification
| Variability Source | Impact Level | Reported Coefficient of Variation | Key Contributing Factors |
|---|---|---|---|
| Data Analysis Software | High | Significant interlaboratory differences | Proprietary instrument software, different calculation algorithms [66] |
| Biological Response | High | >100% (between identically treated individuals) | Genetic causation, physiological differences among test subjects [66] [67] |
| Inter-Assay Technical | Medium | 21.7% | Reagent batches, calibration differences, operator technique [67] |
| Intra-Assay Technical | Low | 11.9% | Pipetting precision, plate effects, reaction efficiency [67] |
| Sample Processing | Medium | Not quantified | RNA extraction methods, reverse transcription efficiency, sample storage [66] |
Performance Comparison of Quantification Methods
Table 2: Performance Metrics of Vtg QPCR Assay Under Standardized Conditions
| Performance Metric | Standardized Protocol Result | Traditional Laboratory-Specific Protocol Result |
|---|---|---|
| Detection Sensitivity | 5.0 ng EE2/L within 24 hours [67] | Variable between laboratories |
| Discriminatory Power | Able to discriminate 10.0 and 5.0 ng EE2/L within 48 hours [67] | Inconsistent discriminatory thresholds |
| Interlaboratory Comparability | High with standardized data analysis [66] | Low, with significant discrepancies |
| Biological Variability Impact | Still present but quantifiable [67] | Confounded with technical variability |
| Transferability Between Labs | Readily transferable [67] | Requires extensive optimization at each site |
Experimental Protocols for Minimizing Variability
Standardized QPCR Protocol for Vtg Quantification
The following protocol was validated across multiple laboratories for robust Vtg transcript quantification:
Sample Collection and RNA Extraction: Collect tissue samples (typically liver) from fathead minnows (Pimephales promelas) following controlled exposures. Homogenize tissue in TRIzol reagent and extract total RNA using silica membrane-based columns. Include DNase treatment to eliminate genomic DNA contamination [66] [67].
Reverse Transcription: Use consistent input amounts of RNA (e.g., 1μg) across all samples. Employ anchored oligo(dT) primers for cDNA synthesis to ensure consistent priming of Vtg mRNA. Use a single reverse transcriptase enzyme lot throughout a complete study [66].
QPCR Assay Conditions: Prepare reaction mixtures containing SYBR Green master mix, gene-specific primers, and cDNA template. Run reactions under the following cycling conditions: initial denaturation at 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Include melt curve analysis to verify amplification specificity [67].
Primer Design and Validation: Design primers to amplify a 100-150 bp region of the fathead minnow Vtg gene. Validate primer efficiency using a standard curve of serial cDNA dilutions, accepting only primers with 90-110% efficiency and single peak melt curves [67].
Critical Modification: Standardized Data Analysis Protocol
The most significant source of interlaboratory variability was identified in the data analysis phase rather than the wet-lab procedures [66]:
- Raw Data Reanalysis: Export raw fluorescence data from all participating laboratories' real-time qPCR machines.
- Unified Analysis Platform: Reanalyze all raw data using independent freeware (LinRegPCR) to calculate cycle thresholds and PCR efficiencies independently of proprietary instrument software.
- Standardized Normalization: Apply consistent normalization algorithms using stable reference genes validated for the specific experimental conditions.
- Cross-Lab Calibration: Use interlaboratory control samples to establish calibration curves and correct for residual systematic biases.
This standardized analysis approach successfully eliminated the interlaboratory variability originating from proprietary software systems, with three of four participating laboratories consistently detecting Vtg in fish exposed to 5 ng/L EE2, and all four laboratories detecting significantly increased Vtg levels in fish exposed to wastewater effluent compared with controls [66].
Visualization of Experimental Workflows
Standardized vs. Traditional Vtg Analysis Pathways
Interlaboratory Variability Reduction Strategy
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagent Solutions for Vtg Quantification
| Reagent/ Material | Function in Vtg Quantification | Implementation Consideration |
|---|---|---|
| LinRegPCR Software | Standardized calculation of PCR efficiency and cycle threshold values | Freeware that eliminates proprietary software variability; essential for cross-lab comparisons [66] |
| SYBR Green Master Mix | Fluorescent detection of amplified DNA sequences | Use consistent lot numbers across collaborating laboratories; validate with standard curves [67] |
| DNase Treatment Kit | Removal of genomic DNA contamination from RNA samples | Critical for preventing false positives; must be included in all RNA extraction protocols [66] |
| Anchored Oligo(dT) Primers | cDNA synthesis specifically from mRNA templates | Preferable to random hexamers for Vtg mRNA to ensure consistent reverse transcription [66] |
| Interlaboratory Control Samples | Calibration and normalization across laboratories | Shared reference materials enable correction of systematic biases between labs [66] |
| Stable Reference Genes | Normalization of Vtg expression levels | Must be validated for specific experimental conditions; multiple reference genes recommended [66] |
| N-acetyl-N-methyl-D-Alanine | N-acetyl-N-methyl-D-Alanine|Research Chemical | N-acetyl-N-methyl-D-Alanine for research applications. This compound is for professional laboratory research use only (RUO), not for personal use. |
| 4-Butyl-3-nitrobenzoic acid | 4-Butyl-3-nitrobenzoic Acid|RUO | 4-Butyl-3-nitrobenzoic acid (C11H13NO4) is a chemical intermediate for research use only. It is not for human or animal consumption. |
Minimizing interlaboratory variability in Vtg quantification requires a systematic approach addressing both technical and analytical procedures. Evidence from comparative studies demonstrates that standardized data analysis using freeware like LinRegPCR effectively reduces variability arising from proprietary instrument software [66]. While biological variability in Vtg expression remains inherent to the system, consistent experimental protocols and standardized analytical methods significantly improve reproducibility across laboratories. Implementation of these evidence-based practices enables reliable detection of Vtg transcriptional responses to environmental estrogens at concentrations as low as 5.0 ng EE2/L, supporting robust biomarker applications in fertility and toxicological research [67]. For optimal results, research programs should prioritize harmonization of data analysis methods while acknowledging and accounting for biological variability through appropriate experimental design and statistical power.
Optimizing dsRNA Delivery for Effective Gene Silencing
The validation of gene function, particularly for critical reproductive genes like vitellogenin (Vg), is a cornerstone of modern fertility research. Vitellogenin is a glycolipoprotein that serves as the primary egg-yolk precursor protein in all oviparous animals, essential for oocyte development and embryonic growth [1]. In the eggplant shoot and fruit borer, Leucinodes orbonalis, CRISPR/Cas9-mediated editing of the vitellogenin gene (LoVg) did not affect the total number of eggs laid but significantly impaired egg hatchability, demonstrating its critical role in viable embryogenesis [68]. This finding highlights the importance of precise gene function analysis in reproductive studies.
RNA interference (RNAi) using double-stranded RNA (dsRNA) has emerged as a powerful tool for functional genomics, allowing researchers to selectively silence genes of interest and investigate their roles in biological processes such as fertility [69]. However, the efficacy of RNAi-based approaches depends heavily on the efficient delivery of intact, stable dsRNA into target cells or organisms. Optimizing dsRNA delivery is thus paramount for obtaining reliable, reproducible results in vitellogenin functional validation studies and broader fertility research programs.
This guide provides a comprehensive, evidence-based comparison of current dsRNA delivery methodologies, supported by experimental data and protocols, to assist researchers in selecting the most appropriate approach for their specific experimental needs in reproductive biology.
Understanding the RNAi Mechanism and Key Components
The RNAi pathway is a conserved biological process that utilizes dsRNA molecules to silence gene expression in a sequence-specific manner. Upon introduction into the cell, long dsRNA is cleaved by the Dicer enzyme into small interfering RNAs (siRNAs) of 21–23 nucleotides [69]. These siRNAs are then loaded into the RNA-induced silencing complex (RISC), where the guide strand directs the complex to complementary mRNA targets for cleavage and degradation, preventing protein translation [69].
Table 1: Key Components of the RNAi Pathway and Their Research Applications
| Component | Description | Research Application in Fertility Studies |
|---|---|---|
| dsRNA | Double-stranded RNA precursor (typically 200-500 bp) | Designed to target vitellogenin or other fertility-associated genes |
| Dicer/Dicer-like Enzymes | RNase III enzymes that process dsRNA into siRNAs | Initiates the silencing cascade; efficiency varies by organism |
| siRNAs | Small interfering RNAs (21-23 nt) that guide RISC to target mRNA | Direct mediators of gene silencing; design affects specificity |
| RISC Complex | Multi-protein complex containing Argonaute proteins | Executes mRNA cleavage; loading efficiency determines silencing efficacy |
| Vitellogenin (Vg) mRNA | Target transcript for silencing in fertility research | Successful reduction indicates delivery efficiency and functional validation |
For vitellogenin research, effective delivery of dsRNA targeting Vg transcripts enables researchers to investigate its pleiotropic functions in reproduction, including its role in oocyte development, nutrient transport to developing embryos, and unexpected functions in social insect behavior and longevity [1].
Figure 1: RNA Interference Mechanism Pathway. dsRNA is processed by Dicer into siRNAs, which are loaded into RISC to guide target mRNA cleavage.
Comparative Analysis of dsRNA Delivery Methods
Physical Delivery Methods
Physical methods utilize mechanical or electrical forces to facilitate dsRNA entry into cells. Electroporation applies electrical pulses to create temporary pores in cell membranes, allowing nucleic acids to enter. The Neon Transfection System demonstrates high efficiency across diverse cell types, including challenging primary cells [70]. While effective for in vitro applications, physical methods generally show limited utility for whole-organism approaches in functional genomics studies targeting reproductive tissues.
Chemical-Based Delivery Systems
Chemical transfection reagents form complexes with dsRNA to facilitate cellular uptake through endocytosis. These methods offer practical advantages for in vitro vitellogenin studies in cell culture systems.
Table 2: Comparative Performance of Leading Transfection Reagents for dsRNA Delivery
| Transfection Reagent | Optimal Application | Efficiency | Cell Viability | Key Advantages | Documented Performance |
|---|---|---|---|---|---|
| Lipofectamine RNAiMAX | siRNA/miRNA delivery | Superior | Superior | Specifically formulated for small RNAs; suitable for reverse transfection | Most efficient reagent for siRNA/miRNA delivery [70] |
| Lipofectamine 3000 | DNA, RNA, co-transfection | Superior | Superior | Versatile for wide range of cell types including difficult-to-transfect cells | Superior efficiency and cell viability for wide range of cells [70] |
| Lipofectamine 2000 | DNA, RNA, co-transfection | High | High | Cost-effective for common cell types | High efficiency for common cell types [70] |
Reverse transfection, where cells are transfected while still in suspension prior to plating, has demonstrated significant advantages over traditional pre-plated methods. This approach increases cell exposure to transfection complexes, often yielding higher efficiency while saving an entire day in experimental workflow [71]. For instance, HepG2 cells, traditionally difficult to transfect, showed remarkable improvement with reverse transfection [71].
Nanotechnology-Enhanced Delivery
Nanoparticle-based delivery systems represent a cutting-edge approach for enhancing dsRNA stability and cellular uptake. Lipid nanoparticles (LNPs), lipoplexes, and polyplexes encapsulate dsRNA, protecting it from degradation and improving delivery efficiency [72] [73]. These nano-delivery systems are particularly valuable for in vivo applications, as they can be engineered for tissue-specific targeting and enhanced endosomal escape.
Recent advances in Spray-Induced Gene Silencing (SIGS) combine dsRNA with nanocarriers to create topical applications that can silence genes in pests and pathogens. This approach has shown promise for agricultural applications but also presents potential for whole-organism studies in insect fertility research [73].
Viral Vector Systems
Viral vectors offer highly efficient delivery for challenging in vivo applications but present significant practical constraints for dsRNA delivery.
Table 3: Viral Vector Systems for Nucleic Acid Delivery
| Vector Type | Payload Capacity | Integration Status | Advantages | Disadvantages | Suitability for Fertility Research |
|---|---|---|---|---|---|
| Adeno-associated Viral Vectors (AAVs) | ~4.7kb | Non-integrating | Mild immune response; FDA approval for some applications | Limited cargo capacity too small for some Cas proteins | Moderate (size limitations may constrain experimental design) |
| Adenoviral Vectors (AdVs) | Up to 36kb | Non-integrating | Large cargo capacity; infects dividing and non-dividing cells | Potential immune responses; off-target effects | High for large genetic constructs |
| Lentiviral Vectors (LVs) | Any size | Integrating | Pseudotyping possible; infects dividing and non-dividing cells | Integration safety concerns; HIV backbone | Moderate (integration may confound fertility studies) |
| Virus-like Particles (VLPs) | Limited | Non-integrating | Reduced safety concerns; transient expression | Manufacturing challenges; stability issues | Emerging technology with potential |
Experimental Protocols for Optimized dsRNA Delivery
Protocol 1: Reverse Transfection for Cell Culture Studies
This protocol adapts methodology from Thermo Fisher Scientific with specific optimizations for fertility research applications [71]:
Day 1: Preparation
- Design and synthesize dsRNA targeting vitellogenin gene sequences (typically 200-500 bp)
- Validate dsRNA quality and concentration via spectrophotometry and gel electrophoresis
- Culture appropriate cell line (e.g., hepatocytes for Vg studies in fish models, insect cell lines for pest fertility research)
Day 2: Reverse Transfection
- Trypsinize and count cells
- Prepare transfection complexes in serum-free medium:
- Dilute 5-50 nM dsRNA in serum-free medium
- Mix with appropriate volume of transfection reagent (e.g., Lipofectamine RNAiMAX)
- Incubate 10-20 minutes at room temperature
- Combine transfection complexes with cell suspension at optimal density (e.g., 1-5×10ⴠcells/cm²)
- Plate cell-transfection complex mixture directly onto culture vessels
- Incubate at standard culture conditions (4-24 hours)
Day 3: Media Replacement and Analysis
- Replace transfection medium with fresh complete medium (if needed based on cytotoxicity)
- Continue incubation until analysis (typically 48-72 hours post-transfection)
- Assess silencing efficiency via qRT-PCR for Vg mRNA and Western blot for Vg protein
Critical Optimization Parameters:
- Cell density: Reverse transfection shows higher tolerance to cell plating density variations compared to traditional methods [71]
- Transfection reagent volume: Must be optimized to balance efficiency and cytotoxicity (e.g., 0.03-1.0 μL/well in 96-well plates) [71]
- Exposure time: Typically 4-24 hours; longer exposure may increase silencing but reduce viability [71]
Protocol 2: dsRNA Sequence Design for Enhanced Efficacy
Optimal dsRNA design significantly improves silencing efficiency, particularly for challenging targets like vitellogenin:
- Target Selection: Identify accessible regions within the vitellogenin mRNA transcript
- Length Optimization: Design dsRNA fragments of 200-500 bp for pesticidal applications or 21-23 nt siRNAs for cell culture [74] [73]
- Sequence Optimization:
- Incorporate thermodynamic asymmetry in siRNA design to favor guide strand loading into RISC [74]
- Select sequences with adenine at the 10th position in antisense siRNA [74]
- Consider higher GC content (9th-14th nucleotides) for insect systems, contrary to mammalian design rules [74]
- Avoid stable secondary structures that may impede processing [74]
- Validation: Use the dsRIP web platform or similar tools to predict efficacy and minimize off-target effects [74]
Figure 2: dsRNA Delivery Experimental Workflow from gene selection to functional analysis.
The Scientist's Toolkit: Essential Research Reagents
Table 4: Essential Research Reagents for Optimized dsRNA Delivery
| Reagent/Category | Specific Examples | Function | Application Notes |
|---|---|---|---|
| Transfection Reagents | Lipofectamine RNAiMAX, Lipofectamine 3000, siPORT NeoFX | Form complexes with dsRNA to facilitate cellular uptake | Select based on cell type; RNAiMAX specifically formulated for RNAi [70] |
| Control RNAs | Silencer GAPDH siRNA, Negative Control #1 siRNA | Transfection efficiency controls and experimental normalization | Essential for distinguishing sequence-specific effects from non-specific responses [71] |
| Cell Viability Assays | ViaCount Assay, MTT, ATP-based assays | Assess cytotoxicity of transfection conditions | Critical for optimizing transfection parameters [71] |
| dsRNA Production Systems | T7 RiboMAX Express RNAi System, in vitro transcription kits | Generate high-quality dsRNA for silencing | Ensure proper length and purity; avoid RNase contamination [70] |
| Detection Reagents | qRT-PCR kits, Western blot reagents, antibodies against Vg | Measure silencing efficiency at mRNA and protein levels | Vg protein detection confirms functional silencing beyond mRNA reduction |
Effective dsRNA delivery for vitellogenin gene silencing requires a multifaceted approach that integrates optimized sequence design with appropriate delivery methodology. For in vitro studies in established cell lines, chemical transfection using reagents like Lipofectamine RNAiMAX with reverse transfection protocols typically provides the most practical balance of efficiency and viability. For challenging in vivo applications or difficult-to-transfect primary cells, nanoparticle-based systems or viral vectors may be necessary despite their additional complexity.
The validation of vitellogenin gene functions in fertility research particularly benefits from approaches that ensure consistent, durable silencing to observe phenotypic effects on oocyte development and embryonic viability. As demonstrated in the Leucinodes orbonalis study, even partial suppression of vitellogenin can significantly impact hatchability without affecting egg production [68], highlighting the importance of delivery optimization to achieve meaningful biological outcomes.
By implementing the comparative data and optimized protocols outlined in this guide, researchers can advance their functional genomics studies with greater confidence in the reliability and interpretability of their experimental results in reproductive biology.
Addressing Species-Specific Considerations in Experimental Design
The vitellogenin (Vtg) gene, which codes for a yolk precursor protein, is a cornerstone for investigating fertility and reproductive biology across oviparous species. However, functional validation of Vtg is highly dependent on the biological context, including taxonomy, reproductive strategy, and social structure. This guide provides an objective comparison of experimental approaches for validating Vtg gene functions, synthesizing current methodological data to aid researchers in selecting and designing robust species-appropriate experiments. A comprehensive understanding of these considerations is critical for advancing research in developmental biology, environmental toxicology, and genetic pest management.
Vitellogenin Functional Validation Across Species: A Comparative Analysis
Experimental data from recent studies reveal how Vtg disruption manifests in distinct phenotypes depending on the species and the technique employed. The following table summarizes key experimental outcomes.
Table 1: Comparative Outcomes of Vitellogenin Gene Disruption Across Species
| Species | Experimental Technique | Key Phenotypic Outcome on Fertility | Quantitative Impact | Citation |
|---|---|---|---|---|
| Tuta absoluta (Tomato Leaf Miner) | RNA interference (RNAi) | Severely impaired ovarian development; reduced fecundity | Shorter ovarian tubes; fewer oocytes; reduced egg hatchability | [17] |
| Leucinodes orbonalis (Eggplant Shoot Borer) | CRISPR/Cas9 knockout | No effect on egg-laying number; significantly reduced egg hatchability | Hatchability negatively affected; total eggs laid unchanged | [68] |
| Apis mellifera (Honey Bee) | RNAi in adult workers | Altered foraging behavior; increased gustatory responsiveness | 96% penetrance of mutant phenotype post-injection | [75] [76] |
| Exopalaemon carinicauda (Ridgetail White Shrimp) | Eyestalk Ablation (induces endogenous Vtg) | Upregulated Vtg synthesis; promoted ovarian development | Significant upregulation of multiple EcVtg mRNAs | [77] |
The data demonstrates that the functional consequences of Vtg manipulation are species-specific. In Lepidoptera like Tuta absoluta, Vg is crucial for ovarian development itself [17]. In contrast, CRISPR/Cas9-mediated knockout in Leucinodes orbonalis suggests Vg is dispensable for oogenesis but essential for embryonic development, as it primarily affects egg hatchability [68]. In the eusocial honey bee, Vg in sterile workers has been co-opted for regulating behavior and lifespan, illustrating a non-reproductive function [76] [78].
Detailed Experimental Protocols for Key Assays
RNAi-Mediated Gene Silencing
This protocol is adapted from successful applications in insects like Tuta absoluta and Apis mellifera [17] [75].
- dsRNA Preparation: Design and synthesize a double-stranded RNA (dsRNA) molecule targeting a 500-600 bp region of the target Vtg mRNA sequence. Use in vitro transcription kits from manufacturers, followed by purification and confirmation via gel electrophoresis.
- Delivery:
- Intra-abdominal Injection (Adult Insects): Anesthetize adults and inject a calibrated dose of dsRNA (e.g., 1-5 µg in honey bees) directly into the hemocoel using a fine glass needle. This method achieved a 96% penetrance in honey bees [75].
- Egg Injection (Embryonic Stage): Inject dsRNA into pre-blastoderm eggs for systemic gene disruption throughout development, though with lower penetrance than adult injection [75].
- Control Groups: Include two control groups: one untreated and one injected with a non-targeting dsRNA (e.g., targeting GFP).
- Phenotypic Assessment:
- Molecular Validation: Quantify Vtg mRNA levels in the fat body and/or hemolymph using qPCR 3-5 days post-injection.
- Physiological Assessment: Dissect ovaries and quantify fecundity (number of eggs laid) and fertility (hatch rate) [17].
- Data Analysis: Compare Vtg expression and reproductive metrics between treatment and control groups using appropriate statistical tests (e.g., t-test, ANOVA).
CRISPR/Cas9 Genome Editing
This protocol is based on the validation of Vg function in Leucinodes orbonalis [68].
- Target Design: Identify a specific 20-nt target sequence within the first exons of the Vg gene. Design single-guide RNAs (sgRNAs) with high on-target efficiency and minimal off-target effects.
- Reagent Delivery: Microinject a mixture of Cas9 protein and sgRNA into early embryos. The optimal concentration ratio must be determined empirically for each species.
- Establishing Mutant Lines: Raise the injected embryos (G0) to adulthood. Outcross individual G0 adults to wild-type partners and screen their progeny (G1) for germline mutations using PCR and sequencing.
- Phenotypic Screening: Cross homozygous G1 or G2 mutants and rigorously assess reproductive phenotypes:
- Count the total number of eggs laid.
- Monitor and calculate the egg hatch rate.
- Examine ovarian development histologically.
- Data Analysis: Compare the fecundity and fertility of mutant lines against wild-type controls. The study on L. orbonalis provides a key reference phenotype where egg-laying is unaffected, but hatchability is severely compromised [68].
Expression Profiling During Ovarian Development
This method is used to correlate Vg expression with reproductive status, as demonstrated in crustaceans and insects [17] [77].
- Sample Collection: Collect target tissues (fat body, hepatopancreas, ovary) from females at different, well-defined stages of ovarian development.
- RNA Extraction and cDNA Synthesis: Homogenize tissues, extract total RNA, and assess quality. Synthesize high-quality cDNA.
- Quantitative PCR (qPCR): Perform qPCR using gene-specific primers for the Vg gene(s). Use at least two validated reference genes (e.g., actin, GAPDH) for normalization.
- Data Analysis: Calculate relative gene expression levels using the 2^(-ΔΔCt) method. Correlate expression dynamics with morphological changes in the ovary.
Visualizing Experimental Workflows and Molecular Relationships
The diagrams below illustrate the core experimental workflow and a key molecular pathway regulating Vg.
Vitellogenin Functional Validation Workflow
Regulatory Feedback Loop in Honey Bees
The Scientist's Toolkit: Key Research Reagent Solutions
Successful investigation of Vtg gene function relies on a suite of specific reagents and tools. The following table details essential materials and their functions.
Table 2: Essential Research Reagents for Vitellogenin Studies
| Reagent / Tool | Primary Function in Experiment | Key Application Notes |
|---|---|---|
| Double-stranded RNA (dsRNA) | Triggers RNAi pathway; degrades complementary Vg mRNA. | Must be designed against a unique region of the Vg transcript; effective in adult honey bees and Lepidoptera [17] [75]. |
| CRISPR/Cas9 System | Creates targeted double-strand breaks in the Vg genomic locus. | Comprises Cas9 nuclease and target-specific sgRNA; can generate non-functional mutants, affecting hatchability without altering fecundity in some pests [68]. |
| Vtg-specific Antibodies | Detects and quantifies Vtg protein levels (via ELISA, Western Blot). | Used for biomarker validation in ecotoxicology; confirms estrogenic exposure in male fish [9] [79]. |
| qPCR Primers/Probes | Quantifies Vg mRNA expression levels in different tissues and stages. | Critical for expression profiling; requires careful validation and normalization to reference genes [17] [77]. |
| Estrogenic Compounds (e.g., EE2) | Positive control for inducing Vg expression in aquatic toxicology. | Used in fish biomarker studies; induces Vtg at concentrations as low as 10 ng/L [9] [79]. |
Quality Control Measures for RNA Integrity and PCR Efficiency
In fertility research, particularly in studies investigating vitellogenin gene functions, reliable gene expression data is paramount. Vitellogenins, the principal yolk proteins in oviparous animals, are essential for reproductive success and serve as critical biomarkers in fertility studies [4] [1]. The validation of their gene functions hinges on accurate molecular analyses, where RNA integrity and PCR efficiency become foundational parameters. RNA quality directly impacts the reproducibility and reliability of gene expression data, while PCR efficiency determines the accuracy of quantification [80] [81]. This guide provides a comparative analysis of current quality control methodologies, experimental protocols, and practical solutions to ensure data integrity in vitellogenin and broader fertility research.
RNA Integrity Assessment: Methods and Comparison
The Importance of RNA Quality in Gene Expression Studies
RNA integrity is the first critical checkpoint in any gene expression workflow. Degraded RNA can severely compromise downstream applications, leading to inaccurate quantification and potentially erroneous biological conclusions [80] [82]. In vitellogenin research, where samples may be derived from various tissues (intestine, liver, fat body) or limited precious specimens, establishing robust RNA quality control is particularly crucial [4] [83]. The vulnerability of RNA molecules to degradation necessitates systematic assessment before proceeding to resource-intensive molecular applications.
Comparative Analysis of RNA Quality Assessment Methods
Table 1: Comparison of RNA Integrity Assessment Methods
| Method | Principle | Output Metrics | Sample Throughput | Cost Considerations | Best Use Cases |
|---|---|---|---|---|---|
| Automated Capillary Electrophoresis | Microfluidic separation and fluorescence detection | RNA Integrity Number (RIN), DV200 | Medium to High | High equipment cost; Moderate per-sample cost | Gold standard for initial quality control; Precise quantification |
| Spectrophotometry | UV absorbance at 260nm, 280nm, and 230nm | A260/A280, A260/A230 ratios, Concentration | High | Low equipment and per-sample cost | Initial purity check; Protein (A260/A280) and solvent contamination (A260/A230) detection |
| 3':5' Assay (qPCR-based) | Amplification efficiency comparison between 3' and 5' ends of transcripts | 3':5' Ratio | Medium | Low to medium per-sample cost | mRNA-specific integrity; When electrophoresis is unavailable |
| Agarose Gel Electrophoresis | Visual assessment of ribosomal RNA bands | 28S:18S ratio, Degradation pattern | Low | Very low | Quick qualitative assessment; When other methods are inaccessible |
RNA Integrity Number (RIN) as a Standard Metric
The RNA Integrity Number (RIN) has emerged as an industry standard for quantifying RNA quality, ranging from 1 (completely degraded) to 10 (perfectly intact) [84]. This algorithm-based metric evaluates the entire electrophoretic trace of an RNA sample, not just the ribosomal ratios, providing a more comprehensive assessment [80]. For downstream applications, RIN values above 8.0 indicate intact RNA highly suitable for sensitive applications, while values between 5.0 and 8.0 represent moderately degraded samples that may still be usable for some applications [82]. Samples with RIN below 5.0 are generally considered too degraded for reliable gene expression analysis [80] [82].
PCR Efficiency Evaluation: Foundations and Methodologies
The Impact of PCR Efficiency on Quantification Accuracy
PCR efficiency represents the amplification success rate per cycle during PCR, ideally approaching 100% (doubling each cycle) [85]. In practical terms, this translates to a slope of -3.32 in a standard curve plot. Deviations from perfect efficiency significantly impact quantification accuracy, especially when using comparative methods like ΔΔCT [85]. The effect becomes particularly pronounced when comparing genes with different amplification efficiencies or when analyzing low-abundance targets like specific vitellogenin isoforms.
Methods for Determining PCR Efficiency
Table 2: PCR Efficiency Determination Methods
| Method | Procedure | Key Output | Advantages | Limitations |
|---|---|---|---|---|
| Standard Curve Method | Serial dilution of template across 3-5 orders of magnitude | Slope, Efficiency (E=10^(-1/slope)-1), R² | Direct measurement; Identifies dynamic range | Requires substantial template; Assumes dilution accuracy |
| Comparative ΔΔCT Validation | Testing amplification of target and reference genes at different template concentrations | Difference in CT values across dilutions | Confirms compatibility for ΔΔCT analysis; No absolute quantification needed | Only confirms comparable efficiency, not exact value |
| Single-Run Efficiency Algorithms | Analysis of amplification curve characteristics | Efficiency without standard curve | Template-saving; Incorporates inhibition detection | Higher variability; Platform-dependent |
Efficiency-Corrected Quantification Models
When PCR efficiencies between target and reference genes differ significantly, efficiency-corrected models should replace the standard ΔΔCT method [81] [85]. The error introduced by efficiency differences can be substantial, with a PCR efficiency of 0.9 (instead of 1.0) resulting in a 261% error at a threshold cycle of 25 [85]. This miscalculation could lead to a 3.6-fold underestimation of actual expression levels, critically impacting conclusions about vitellogenin gene regulation in fertility studies.
Integrated Experimental Protocols
Protocol 1: Comprehensive RNA Quality Assessment
Principle: Evaluate RNA samples through complementary methods to confirm integrity, purity, and concentration.
Materials:
- QIAxcel Advanced system (Qiagen) or Bioanalyzer (Agilent)
- Nanodrop or similar spectrophotometer
- RNA 6000 Nano Kit (Agilent)
- Nuclease-free water and tubes
Procedure:
- Spectrophotometric Analysis:
- Use 1-2μL RNA sample for A260/A280 and A260/A230 ratios.
- Acceptable ranges: A260/A280 ≈ 2.0 (1.8-2.1); A260/A230 > 1.8 [84].
- Record concentration for subsequent dilutions.
Capillary Electrophoresis:
- Prepare samples according to manufacturer's protocol (typically 1μL RNA).
- Run on appropriate platform to generate electropherograms and RIN values.
- Visually inspect electrophoretic traces for abnormal profiles.
qPCR-based 3':5' Assay (Alternative/Complementary):
- Design primer sets targeting 3' and 5' regions of a stable housekeeping gene (e.g., Pgk1) [82].
- Perform RT-qPCR with both primer sets using the same cDNA synthesis reaction.
- Calculate 3':5' ratio; values approaching 1.0 indicate intact mRNA.
Troubleshooting: Low A260/A230 suggests solvent carryover; low RIN with good ratios indicates ribosomal RNA preservation but potential mRNA degradation; elevated 3':5' ratios (>2.0) confirm mRNA degradation.
Protocol 2: PCR Efficiency Determination and Validation
Principle: Establish amplification efficiency for each primer set to guide quantification model selection.
Materials:
- Validated RNA samples (RIN > 7.0)
- Reverse transcription kit (e.g., Qiagen Quantitect)
- qPCR master mix (SYBR Green or probe-based)
- Primers for target genes (vitellogenin isoforms) and reference genes
Procedure:
- Standard Curve Preparation:
- Perform serial dilutions (minimum 5 points, 10-fold or 5-fold) of pooled cDNA.
- Include triplicate reactions for each dilution point.
- Run qPCR program with appropriate cycling conditions.
Data Analysis:
- Plot CT values against log template concentration.
- Calculate slope from linear regression: Efficiency = 10^(-1/slope) - 1 [85].
- Acceptable efficiency range: 90-110% (slope -3.6 to -3.1).
Comparative Efficiency Validation:
- For ΔΔCT applications, test target and reference genes with the same dilution series.
- Plot ΔCT (CTtarget - CTreference) against log dilution.
- Slope < 0.1 indicates sufficiently similar efficiencies for ΔΔCT method [85].
Primer Optimization Considerations: Test primer concentrations (50-800 nM), annealing temperature gradients (55-65°C), and validate specificity via melt curve analysis or gel electrophoresis [86].
Visualizing Quality Control Workflows
Diagram 1: Integrated workflow for RNA quality control and PCR efficiency validation in gene expression studies, highlighting critical checkpoints for reliable vitellogenin gene analysis.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Essential Reagents for RNA Quality Control and PCR Validation
| Reagent/Category | Specific Examples | Primary Function | Application Notes |
|---|---|---|---|
| RNA Extraction Kits | RNeasy Mini Plus (Qiagen), TRIzol | High-quality RNA purification with genomic DNA removal | Column-based methods preferred for consistent yield and purity |
| RNA Quality Assessment | RNA 6000 Nano Kit (Agilent), QIAxcel RNA Kit | RIN determination and integrity verification | Essential pre-screening step; correlates with qPCR performance |
| Reverse Transcription | Quantitect Reverse Transcription (Qiagen), High-Capacity cDNA | cDNA synthesis with controlled RNA input | Use consistent input (100-500ng) across samples; include no-RT controls |
| qPCR Master Mixes | SYBR Green, TaqMan assays | Specific and sensitive detection | SYBR Green requires optimization; TaqMan offers higher specificity |
| Reference Genes | Pgk1, Actb, Gapdh, 18S rRNA | Expression normalization | Must be validated for specific tissue and experimental conditions |
| Efficiency Standards | Serial dilution templates, Synthetic oligonucleotides | Standard curve generation | Use pooled cDNA for biologically relevant efficiency measures |
| Primer Validation Tools | OligoArchitect, Primer-BLAST | In silico specificity and dimer analysis | Critical step to minimize off-target amplification and primer dimers |
Quality control measures for RNA integrity and PCR efficiency are not merely optional precautions but fundamental requirements for robust gene expression analysis, particularly in complex studies such as vitellogenin gene function validation in fertility research. The integrated approach presented here—combining multiple RNA assessment methods with rigorous PCR validation—provides a framework for generating reliable, reproducible data. As research progresses toward increasingly subtle biological questions, from vitellogenin isoform-specific functions to intergenerational signaling effects [4] [83], the implementation of comprehensive QC protocols becomes ever more critical for scientific advancement and discovery validation.
In the study of fertility biomarkers such as vitellogenin (Vg), the journey from raw fluorescence measurements to biologically meaningful expression data is a critical methodological pathway. Vitellogenin, a glycolipoprotein essential for oocyte development and reproductive regulation, serves as a crucial biomarker in fertility studies across insect species and other oviparous animals [1]. The accurate quantification of its expression levels, often through fluorescence-based techniques like flow cytometry, is fundamental to understanding reproductive biology and developing potential interventions for pest control or fertility enhancement. However, technical variations in instrumentation, reagent lots, and sample handling can introduce significant artifacts that obscure true biological signals, particularly when comparing across multiple samples or experimental batches [87] [88]. This challenge is especially pronounced in large-scale studies such as clinical trials or multi-center research initiatives where subtle biological differences must be distinguished amidst substantial technical noise.
Normalization of fluorescence data addresses these challenges by systematically removing technical between-sample variation while preserving biologically relevant information. For researchers investigating vitellogenin gene functions in fertility, employing robust normalization practices is not merely an optional preprocessing step but a fundamental requirement for generating reliable, reproducible conclusions about gene expression, protein quantification, and their correlation with reproductive outcomes. The following sections provide a comprehensive framework for navigating this analytical pipeline, from experimental design through data interpretation, with particular emphasis on methodologies validated in reproductive biology contexts.
Experimental Protocols: Establishing a Foundation for Quality Data
Flow Cytometry and Fluorescence Detection Fundamentals
Fluorescence detectors operate on the principle of measuring light emitted by fluorophores after excitation at a specific wavelength. The process involves absorption of light by a fluorophore, which then emits light at a longer wavelength as it returns to its ground state. The emitted fluorescence intensity (If) is theoretically described by the equation: If = ε × c × Φf × I0, where ε is the molar absorptivity, c is the concentration of the fluorophore, Φf is the fluorescence quantum yield, and I0 is the excitation light intensity [89]. This relationship forms the theoretical foundation for quantitative analysis in fluorescence-based applications, including vitellogenin detection.
Successful fluorescence detection begins with appropriate instrument selection based on experimental needs. Spectrofluorometers, which measure complete fluorescence emission spectra, are ideal for detailed characterization of fluorophore properties, while filter fluorometers, utilizing specific wavelength filters, offer robust solutions for quantitative analysis of known fluorophores like those used in vitellogenin tagging [89]. For cellular localization studies of vitellogenin expression, fluorescence microscopes provide spatial resolution necessary for determining tissue-specific expression patterns in ovaries or fat bodies [89].
Controls and Experimental Design for Vitellogenin Studies
Incorporating appropriate controls is essential for meaningful interpretation of vitellogenin expression data. Experimental designs should include:
- Isotype controls to account for non-specific antibody binding [90]
- Negative controls from tissues or cells not expressing vitellogenin (e.g., male specimens where Vg is typically absent) to establish baseline fluorescence [1]
- Positive controls with known vitellogenin expression levels for normalization across experiments
- Reference standards for instrument calibration and quantitative comparison across experimental batches
For vitellogenin functional studies, particularly those employing RNAi-mediated gene silencing, experimental designs should account for the temporal dynamics of gene expression reduction and its phenotypic manifestations. As demonstrated in Tuta absoluta research, effective TaVg silencing requires monitoring throughout ovarian development stages, with phenotypic assessments including ovarian tube length measurements, oocyte counts, yolk deposition quantification, and egg viability tests [17].
Sample Preparation Protocols
Proper sample preparation minimizes technical variation and enhances normalization effectiveness:
- Tissue processing: For vitellogenin studies, fat body and ovarian tissues require careful dissection and homogenization to preserve protein integrity [91] [17]
- Fixation considerations: When using intracellular staining for vitellogenin detection, fixation methods must balance epitope preservation with membrane permeability
- Concentration optimization: Sample concentration should be adjusted to avoid self-quenching or inner filter effects that non-linearly affect fluorescence intensity [89]
- Solvent compatibility: Use solvents that minimize background fluorescence while maintaining fluorophore stability [89]
Normalization Methodologies: From Global to Local Approaches
Theoretical Framework for Normalization
Normalization addresses technical variation in high-throughput fluorescence data by aligning prominent features (landmarks) across samples. In flow cytometry and other fluorescence-based applications, this technical variation manifests as shifts in the absolute position of cell populations or expression peaks when displayed on a common axis, complicating biological interpretation [87]. These technical variations arise from multiple sources, including changes in instrumentation channel voltages, switching antibody variants, reagent lot differences, and instrument drift over time [88].
The fundamental principle underlying normalization is the identification of stable biological features that should remain constant across samples, then applying mathematical transformations to align these features while preserving biologically relevant differences. For vitellogenin expression studies, this might involve identifying invariant housekeeping genes or stable cell populations that serve as reference points for technical variation correction.
Implementation of Normalization Algorithms
Two prominent normalization methodologies with particular relevance to fluorescence data are the gaussNorm and fdaNorm algorithms, both available through the Bioconductor flowStats package [87]. These methods share a common three-step framework but differ in their implementation:
Landmark Identification: This critical first step identifies peaks in kernel density estimates that correspond to biologically stable cell populations or expression values. The gaussNorm approach identifies local maxima in kernel density estimates and applies a confidence score based on both peak sharpness and height to distinguish true biological populations from spurious peaks [87]. The fdaNorm method employs a robust statistical testing framework to infer significant landmarks based on gradient and curvature derivatives of modal regions, typically yielding fewer spurious peaks [87].
Landmark Registration: This step establishes correspondence between peaks identified in individual samples and a reference set of landmarks. When the number of peaks matches the number of reference landmarks, labeling proceeds consecutively with respect to location. For mismatched populations, gaussNorm uses a median-based matching algorithm while fdaNorm employs k-means clustering to assign landmark labels [87].
Landmark Alignment: The final step transforms sample data so that identified landmarks align with their reference positions. gaussNorm uses an exponential decay function where data point shifting decreases as distance from landmarks increases, allowing independent adjustment of landmark positions [87]. fdaNorm represents kernel density estimates with B-spline interpolands for transformation.
Local vs. Global Normalization
A significant advancement in normalization practice is the shift from global to local normalization approaches. Global normalization applies the same transformation to all cells in a channel, while local normalization integrates with the gating structure to apply transformations specific to defined cell subsets [88]. This approach is particularly valuable for vitellogenin studies where expression may be tissue-specific (e.g., fat body versus ovarian expression).
Local normalization demonstrates superior performance in clinical trial data sets, as evidenced by applications in HIV Vaccine Trials Network (HVTN) and Immune Tolerance Network (ITN) studies. In B-cell phenotyping data, local normalization combined with template gating performed as well as manual gating with individual adjustment for each sample, while significantly reducing analyst time and subjective bias [88]. In intracellular cytokine staining assays, local normalization effectively mitigated false positive response calls that occurred with global approaches [88].
Table 1: Comparison of Normalization Methods for Fluorescence Data
| Method | Algorithm Type | Key Features | Best Applications | Limitations |
|---|---|---|---|---|
| gaussNorm | Peak-based alignment | Uses confidence scores for peak selection; exponential decay transformation | Studies with well-defined cell populations; when number of landmarks is known | Struggles with overlapping populations; sensitive to bandwidth selection |
| fdaNorm | Functional data analysis | Uses statistical testing for landmarks; B-spline transformations | Data with subtle population differences; automated landmark number detection | Computational intensity; complex implementation |
| Local Normalization | Gating-integrated | Applies normalization within specific cell subsets; template-based | Large clinical trials; studies with predefined cell populations of interest | Requires preliminary gating hierarchy |
| Global Normalization | Channel-wide transformation | Applies same transformation to all cells in a channel | Preliminary analyses; data with minimal population overlap | Can miss population-specific variation; may distort minor subsets |
Data Interpretation and Analytical Workflows
Gating Strategies for Vitellogenin Expression Analysis
Effective interpretation of fluorescence data requires systematic gating strategies to identify cell populations of interest. For vitellogenin research, this typically begins with forward scatter (FSC) versus side scatter (SSC) plots to gate on viable cells, followed by fluorescence-based gating to identify vitellogenin-positive populations [90]. Data representation choices significantly impact interpretation efficacy:
- Histograms: Ideal for single-parameter data visualization, such as vitellogenin fluorescence intensity distribution. Overlaying histograms from different samples (e.g., experimental versus control) allows direct comparison of expression shifts [90]
- Scatter plots: Essential for multiparameter analysis, enabling simultaneous visualization of vitellogenin expression against other cellular markers. Dot plots, density plots, and contour plots offer complementary visualization advantages for identifying cell subpopulations [90]
- Quadrant analysis: Using differential fluorescence thresholds to categorize cells as double-positive, double-negative, or single-positive for multiple markers of interest
Following normalization, the gating process becomes more reproducible and less dependent on individual analyst adjustment. Template gating approaches, where a single gate set is applied across multiple normalized samples, become feasible after effective normalization, significantly accelerating analysis of large data sets [88].
Troubleshooting and Quality Assessment
Robust data interpretation requires systematic quality assessment and troubleshooting protocols. Common issues in fluorescence data analysis include:
- Instrument noise: Addressed through regular calibration, optics cleaning, and component replacement [89]
- Sample degradation: Minimized through optimized sample handling, appropriate solvents, and proper storage conditions [89]
- Optical alignment problems: Corrected through instrumental maintenance and alignment verification [89]
- Background fluorescence: Managed through solvent selection and appropriate control samples
Quality metrics for normalization effectiveness include population stability (assessed through coefficient of variation across replicates), signal-to-noise ratios, and concordance between technical replicates. For vitellogenin studies specifically, quality assessment should include verification of expected expression patterns (e.g., female-specific expression, higher expression in reproductive tissues) [1].
Application to Vitellogenin Functional Validation
Vitellogenin Biology and Research Significance
Vitellogenin represents a critical research target in fertility studies due to its fundamental role in reproductive biology across oviparous species. As a glycolipoprotein belonging to the large lipid transport protein family, vitellogenin functions in transporting maternal lipids, carbohydrates, metals, and phosphorous to developing oocytes [1]. Its expression is predominantly female-specific in most species, with any detectable expression in males suggesting potential endocrine disruption by environmental contaminants [1].
Recent research has revealed vitellogenin's pleiotropic functions beyond reproduction, including roles in oxidative stress protection, immunomodulation, and social behavior regulation in eusocial insects like honeybees [1]. This functional diversity, coupled with its essential reproductive role, makes vitellogenin an compelling target for both basic research and applied pest management strategies. In the red imported fire ant (Solenopsis invicta), comparative analyses of reproductive caste types have identified specific vitellogenin genes (Vg2 and Vg3) involved in queen fertility, with knockdown experiments demonstrating significant impacts on oogenesis and egg production [91].
Experimental Validation of Vitellogenin Function
Functional validation of vitellogenin in fertility typically combines gene expression analysis with phenotypic assessment. In Tuta absoluta, RNAi-mediated silencing of TaVg resulted in significant downregulation of vitellogenin content, severely affecting ovarian development with shorter ovarian tubes, reduced oocyte counts, decreased yolk deposition in egg chambers, and significantly reduced egg production and hatching rates [17]. Similar approaches in Solenopsis invicta demonstrated that downregulation of SiVg2 and SiVg3 led to smaller ovaries, impaired oogenesis, and reduced egg production [91].
These functional studies rely heavily on robust fluorescence-based quantification methods. Normalization ensures that observed expression differences reflect biological reality rather than technical artifacts, particularly important when comparing across experimental conditions, developmental timepoints, or tissue types. The table below summarizes key experimental findings from recent vitellogenin functional studies.
Table 2: Vitellogenin Functional Validation in Recent Research
| Study System | Experimental Approach | Key Functional Findings | Expression Measurement Method |
|---|---|---|---|
| Solenopsis invicta (Red imported fire ant) [91] | RNAi-mediated gene silencing | Vg2 and Vg3 knockdown reduced ovary size, oogenesis, and egg production; identified as regulators of queen fecundity | RNA-seq, qRT-PCR validation |
| Tuta absoluta (Tomato leaf miner) [17] | RNAi silencing of TaVg and TaVgR | Reduced vitellogenin content, shorter ovarian tubes, fewer oocytes, decreased egg production and hatching | Spatiotemporal expression analysis |
| Honeybee (Apis mellifera) [1] | Vitellogenin pleiotropy studies | Roles in social behavior, lifespan, immunity, and oxidative stress protection beyond reproduction | Protein quantification, functional assays |
| Zebrafish (Danio rerio) [1] | Environmental contaminant exposure | Hepatic vitellogenin induction in males indicating estrogenic activity; impaired reproduction | Plasma protein measurement |
Research Reagent Solutions for Vitellogenin Studies
Table 3: Essential Research Reagents for Vitellogenin Functional Studies
| Reagent Category | Specific Examples | Research Function | Application Notes |
|---|---|---|---|
| Fluorophore-Conjugated Antibodies | Anti-vitellogenin primary antibodies with FITC, PE, APC conjugates [92] | Detection and quantification of vitellogenin protein expression | Validation for specific species required; concentration optimization critical |
| RNAi Reagents | dsRNA targeting Vg genes [91] [17] | Gene silencing for functional validation | Requires sequence-specific design; efficiency monitoring essential |
| qPCR Assays | Sequence-specific primers for Vg genes [91] | Gene expression quantification | Should target conserved regions; require reference gene normalization |
| Flow Cytometry Panel Components | Cell viability dyes, surface marker antibodies [90] | Cell population identification and sorting | Panel design must consider fluorophore spectral overlap |
| Normalization Tools | Bioconductor flowStats package [87] | Computational normalization of fluorescence data | Algorithm selection dependent on data structure and biological question |
Visualizing Analytical Workflows
The following diagram illustrates the integrated workflow from sample preparation through normalized data analysis, highlighting key decision points and quality control checkpoints essential for reliable vitellogenin research.
Figure 1: Analytical Workflow from Sample to Insight
The integration of robust normalization practices into vitellogenin research protocols represents a methodological imperative for generating reliable, reproducible findings. As fluorescence-based technologies continue to advance, enabling increasingly multiplexed experimental designs, the importance of systematic data processing only grows more pronounced. For researchers investigating the role of vitellogenin in fertility, adopting the best practices outlined herein—appropriate experimental design with controls, careful instrument selection and calibration, methodical normalization implementation, and systematic data interpretation—provides a solid foundation for meaningful biological discovery.
The demonstrated role of specific vitellogenin genes in regulating fertility across diverse species, from fire ants to tomato leaf miners, highlights the conservation of vitellogenin function in reproduction and its potential as a target for fertility manipulation. By implementing rigorous data analysis practices from raw fluorescence to normalized expression, researchers can accelerate progress in both fundamental reproductive biology and applied pest management strategies rooted in vitellogenin functional disruption.
Cross-Species Validation Evidence: Linking Vtg Disruption to Fertility Impairment
The red palm weevil, Rhynchophorus ferrugineus, is a globally invasive pest responsible for devastating losses in palm plantations worldwide. This case study examines RNA interference (RNAi) technology targeting the vitellogenin (Vg) gene, a critical regulator of reproduction, as a novel biocontrol strategy. We present a comprehensive analysis of experimental protocols, quantitative results on knockdown efficacy, and comparative performance against alternative RNAi targets and conventional control methods. The data demonstrate that Vg-based RNAi achieves near-complete suppression of oogenesis and egg viability, positioning it as a highly specific and potent tool for population management within an integrated pest management framework.
The Pest: Rhynchophorus ferrugineus
Rhynchophorus ferrugineus (Oliver), commonly known as the red palm weevil (RPW), is a highly destructive pest native to South and Southeast Asia that has rapidly expanded its geographical range to most palm-growing regions globally [93] [94]. This insect poses a severe threat to ornamental and commercial palm species, particularly date palms (Phoenix dactylifera) and coconut palms (Cocos nucifera), with a host range encompassing over 40 palm species [93] [94]. The weevil's cryptic nature, with larvae completing their entire development concealed inside palm trunks, complicates early detection and limits the efficacy of conventional control methods, including chemical insecticides [95] [94]. Infested palms typically show visible symptoms only after extensive internal damage has occurred, often leading to tree mortality [94].
The Vitellogenin Gene in Insect Reproduction
Vitellogenin (Vg) is a glycolipoprotein that serves as the major yolk protein precursor in all oviparous organisms, including insects [95] [17]. In female insects, Vg is synthesized in the fat body, secreted into the hemolymph, and selectively taken up by developing oocytes via receptor-mediated endocytosis [95]. Once incorporated into oocytes, it is stored as vitellin (Vn), which provides the primary nutritional reserve for embryonic development [95]. The critical role of Vg in reproduction makes it a promising target for disrupting insect population growth. RNAi-mediated silencing of Vg has been successfully demonstrated to impair ovarian development and reduce fecundity in multiple insect species, including the warehouse moth (Cadra cautella) and the tomato leaf miner (Tuta absoluta) [96] [17].
Experimental Protocols
Gene Characterization and Target Selection
The complete RfVg transcript was sequenced and characterized, revealing a 5504 bp sequence encoding a 1787 amino acid protein [95]. Structural analysis identified conserved domains typical of insect vitellogenins:
- Vitellogenin_N domain (amino acids 21-735)
- Domain of Unknown Function 1943 (DUF1943) (amino acids 769-1059)
- Von Willebrand factor type D domain (VWD) (amino acids 1467-1657) [95]
The expression profile of RfVg was determined through RT-PCR analysis, confirming exclusive expression in female fat body cells with temporal expression beginning on the first day of adulthood and intensifying after emergence from cocoons [95].
dsRNA Design and Synthesis
A double-stranded RNA (dsRNA) targeting a unique 400 bp region (position 3538-3938 bp) of the RfVg transcript was designed [95]. This region showed very low or no homology with other insect Vg genes to ensure target specificity and minimize off-target effects. The dsRNA was synthesized using standard in vitro transcription methods, purified, and resuspended in nuclease-free buffer for injection.
Bioassay and Delivery Methods
The RNAi bioassay employed microinjection as the primary delivery method:
- Adult female weevils received dorsally injected dsRNA at a dose of 2 μg per weevil [95]
- Control groups received injections of nonsense dsRNA or buffer alone
- Treated weevils were maintained under laboratory conditions with appropriate nutrition
- Tissues were collected at 15, 20, and 25 days post-injection for molecular and phenotypic analysis
Assessment Methods
Gene Silencing Efficacy: Quantitative real-time PCR (qRT-PCR) was performed to measure RfVg transcript levels using gene-specific primers, with tubulin serving as the internal reference gene [95].
Protein Analysis: Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to detect reductions in Vg protein expression in fat body and hemolymph samples [95].
Phenotypic Assessment: Ovarian development was evaluated through morphological examination of ovarian tubes, oocyte counts, and measurement of yolk deposition. Fecundity was assessed by counting eggs laid, and hatchability was determined by monitoring egg development [95].
Figure 1: Experimental workflow for RNAi-mediated Vg knockdown in R. ferrugineus.
Results and Comparative Analysis
Efficacy of Vg Gene Silencing
RNAi-mediated targeting of RfVg resulted in significant and progressive suppression of target gene expression. Quantitative analysis revealed a time-dependent increase in knockdown efficiency, achieving near-complete silencing by 25 days post-treatment [95].
Table 1: Time-course quantification of RfVg gene silencing efficacy
| Days Post-injection | Knockdown Efficiency (%) | Residual Expression Level |
|---|---|---|
| 15 | 95.0% | 5.0% |
| 20 | 96.6% | 3.4% |
| 25 | 99.0% | 1.0% |
The dramatic reduction in RfVg transcript levels directly correlated with impaired Vg protein synthesis, as confirmed by SDS-PAGE analysis, which showed failure of Vg protein expression in treated females [95].
Phenotypic Consequences of Vg Knockdown
Suppression of RfVg expression resulted in profound impairment of reproductive function, demonstrating the critical role of vitellogenin in oogenesis and embryonic development [95].
Table 2: Phenotypic effects of RfVg silencing on reproductive parameters
| Reproductive Parameter | dsRNA-Treated Group | Control Group |
|---|---|---|
| Ovarian Development | Atrophied ovaries, no oogenesis | Normal development |
| Oocyte Production | Severely reduced or absent | Normal oogenesis |
| Yolk Deposition | Dramatically reduced | Normal deposition |
| Egg Hatchability | Complete failure (0%) | Normal hatchability |
Female weevils subjected to RfVg knockdown exhibited atrophied ovaries with significantly reduced oocyte production and yolk deposition in egg chambers. While some females retained the capacity to lay eggs, these eggs completely failed to hatch, demonstrating the essential role of Vg in embryonic viability [95].
Comparative Performance Against Alternative RNAi Targets
Catalase gene silencing represents an alternative RNAi approach for R. ferrugineus management, demonstrating insecticidal potency through induction of oxidative stress and mortality across developmental stages [97].
Table 3: Comparative efficacy of different RNAi targets in R. ferrugineus
| Parameter | Vg-Targeted RNAi | Catalase-Targeted RNAi |
|---|---|---|
| Target Gene Function | Reproduction (Yolk protein precursor) | Antioxidant defense (Reactive oxygen species detoxification) |
| Primary Effect | Reproductive failure | Mortality and growth inhibition |
| Efficacy in Larvae | Not reported | 80% mortality (5th instar) |
| Efficacy in Adults | 99% gene knockdown | 30% mortality (10th instar) |
| Time to Maximum Effect | 25 days | Varies by instar |
| Delivery Method | Injection (2μg/weevil) | Injection, artificial diet, topical application |
While catalase knockdown achieved significant mortality in larval stages (80% in fifth instar), Vg targeting offers the strategic advantage of population suppression through reproductive disruption rather than direct mortality, potentially providing longer-term control by preventing population recruitment [95] [97].
Advantages Over Conventional Control Methods
Current RPW management relies heavily on chemical insecticides, which face limitations due to the insect's cryptic behavior, development of resistance, and environmental concerns [95] [94]. Vg-targeted RNAi addresses several key limitations of conventional approaches:
- Specificity: Targets a vital reproductive gene with minimal non-target effects
- Efficacy: Achieves near-complete reproductive disruption
- Environmental Safety: Reduced ecological impact compared to broad-spectrum insecticides
- Sustainability: Functions at low concentrations without reported resistance
Figure 2: Logical pathway of RfVg silencing consequences from molecular to population level.
The Scientist's Toolkit: Research Reagent Solutions
Table 4: Essential research reagents for RNAi-based fertility studies in insects
| Reagent / Material | Function / Application | Specifications / Notes |
|---|---|---|
| Gene-Specific Primers | Amplification and quantification of target gene | Design for unique region (e.g., 3538-3938 bp of RfVg) |
| In Vitro Transcription Kit | dsRNA synthesis | High-yield, nuclease-free production |
| Microinjection System | Precise delivery of dsRNA | Calibrated for 2μL injections in adult weevils |
| qRT-PCR Reagents | Quantification of gene expression | Include reverse transcription and SYBR Green components |
| SDS-PAGE Equipment | Protein analysis and confirmation | Verification of Vg protein knockdown |
| Artificial Diet | Laboratory rearing of insects | Contains agar, yeast, wheat meal, vitamins [93] |
| RNA Stabilization Solution | Preservation of RNA integrity | Immediate tissue preservation post-collection |
RNAi-mediated silencing of the vitellogenin gene represents a highly efficacious and specific approach for population control of Rhynchophorus ferrugineus. The experimental data demonstrate that RfVg knockdown achieves near-complete suppression of reproduction through disruption of oogenesis and egg viability, without direct mortality of adult insects. This case study validates Vg as a critical genetic determinant of fertility in insects and highlights its potential as a target for species-specific biocontrol strategies. While delivery challenges remain for field application, the robust laboratory efficacy provides a compelling foundation for further development of RNAi-based technologies in sustainable pest management.
Temperature Effects on VgR Expression and Reproduction in Zeugodacus cucurbitae
Within the context of global climate change, understanding the molecular mechanisms through which extreme temperatures affect pest fertility is crucial for developing targeted control strategies. This guide objectively compares the roles of key reproductive genes, notably the vitellogenin receptor (VgR), in the melon fly, Zeugodacus cucurbitae, under short-term high-temperature stress. We summarize and contrast experimental data on how silencing VgR and related vitellogenin (Vg) genes disrupts oviposition, ovarian development, and lifespan, providing a validated molecular target for fertility disruption in this significant agricultural pest.
Comparative Analysis of Gene Function in Reproduction Under Heat Stress
The following table summarizes the phenotypic outcomes resulting from the silencing of key reproduction-related genes in Z. cucurbitae under high-temperature conditions.
Table 1: Comparative Impacts of Silencing Reproduction-Related Genes under Short-Term High-Temperature Stress (45°C for 1 hour) in Zeugodacus cucurbitae
| Gene Silenced | Impact on Fecundity (Egg Production) | Impact on Ovary Development | Impact on Adult Lifespan | Key Synergistic Gene Expression Changes |
|---|---|---|---|---|
| VgR (ZcVgR) | Reduction of 95.2% compared to control [98]. | Significant decrease in development speed and ovarian diameter [98]. | Significantly reduced [98]. | Downregulation of HSP70, JHEH, TnC, and ZcVg1 [99]. |
| Vitellogenin-3 (ZcVg3) | Reduction of 84.7% compared to control [100]. | Significantly decreased development rate and ovarian diameter [100]. | Reduced by 71% [100]. | Upregulation of ZcVg1 and ZcVg2; variable effect on ZcVgR [100]. |
The data presented in Table 1 demonstrates that targeting the vitellogenin pathway, particularly through VgR, is highly effective for suppressing reproduction. The extreme sensitivity of this system to temperature is highlighted by the near-total cessation of egg production following ZcVgR knockdown at 45°C.
Experimental Protocols for Functional Gene Validation
A critical methodology for validating the function of these genes involves RNA interference (RNAi) coupled with physiological assays under controlled temperature regimes. The following diagram outlines a standard experimental workflow for this functional validation.
Experimental Workflow for Validating Gene Function in Z. cucurbitae
Insect Rearing and Temperature Treatment
- Insect Source: Z. cucurbitae populations are typically collected from infested fields (e.g., bitter gourd in Hainan Province, China) and maintained in the laboratory for multiple generations [100] [101] [98].
- Standard Rearing Conditions: Insects are reared at 25 ± 1°C, 70 ± 5% relative humidity, with a photoperiod of 14 hours light:10 hours dark [100] [98]. Larvae are fed an artificial diet containing pumpkin, cornflour, yeast, and sucrose, while adults are fed a mixture of sucrose and yeast powder [101] [98].
- Heat Stress Application: Newly emerged adults are transferred to an artificial climate chamber and subjected to a defined short-term high-temperature stress, such as 45°C for 1 hour, with a 25°C group serving as the control [98]. This specific stressor has been shown to stimulate reproduction in contemporary generations, making it a relevant stress model [101] [98].
Gene Silencing via RNA Interference (RNAi)
- siRNA Design: Based on the conserved cDNA sequence of the target gene (e.g.,
ZcVgRorZcVg3), a 19-nucleotide siRNA is designed and synthesized commercially [100]. - dsRNA/siRNA Delivery: The silencing agent is injected directly into the abdomen of sexually mature females (e.g., 5-day-old adults) using a micro-injector [98]. Control groups are typically injected with a non-targeting (e.g., GFP) dsRNA or buffer.
Molecular and Phenotypic Assessment
- Knockdown Efficiency Validation: At specified intervals (e.g., 24 and 72 hours post-injection), total RNA is extracted from female flies. The silencing efficiency is evaluated using Reverse Transcription Quantitative PCR (RT-qPCR) to measure the relative expression level of the target gene compared to controls [98].
- Phenotypic Measurements:
- Fecundity: The total number of eggs laid, oviposition days, and egg hatchability are recorded daily [98].
- Ovarian Development: Ovaries are dissected and examined under a microscope. The development speed and ovarian diameter are measured and compared to controls [100] [98].
- Lifespan: The longevity of treated and control adult flies is tracked and recorded [100] [98].
Signaling Pathways and Molecular Interactions
The molecular response to VgR gene silencing under heat stress involves a complex network of pathways affecting immunity, metabolism, and critical reproductive functions. The diagram below summarizes these key interactions.
Molecular Network Upon ZcVgR Silencing Under Heat Stress
The molecular network triggered by ZcVgR knockdown, as illustrated above, shows a cascade of events leading to the observed phenotypic outcomes.
- Transcriptomic Level: Following
ZcVgRsilencing, genes includingvitellogenin-1 (ZcVg1),juvenile hormone epoxide hydrolase (JHEH),troponin C (TnC), andheat shock protein 70 (HSP70)are significantly downregulated [99]. Concurrently, immune-related pathways are activated, while energy metabolism-related pathways are inhibited [99]. - Proteomic Level: A correlation analysis between transcriptome and proteome revealed a low correlation, indicating complex post-transcriptional regulation. At the protein level, glycometabolism and phagosome pathways are activated, while phototransduction-fly and autophagy-animal pathways are inhibited [99].
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagents and Resources for Functional Studies in Zeugodacus cucurbitae
| Reagent/Resource | Function/Application in Research | Example Source / Specification |
|---|---|---|
| Artificial Diet | Standardized laboratory rearing of all life stages. | Formulations include pumpkin, cornflour, yeast powder, sucrose, and preservatives [101] [98]. |
| siRNA / dsRNA | Gene silencing via RNA interference. | Custom-designed 19-nucleotide sequences targeting conserved regions of genes like ZcVgR [100] [98]. |
| Artificial Climate Chamber | Application of precise, short-term temperature stress. | Enables treatment at defined temperatures (e.g., 25°C-45°C) and humidity [98]. |
| TRIzol Reagent | Isolation of high-quality total RNA from insect tissues. | Used for RNA extraction prior to RT-qPCR or transcriptome sequencing [102]. |
| TMT Proteomics Kit | Quantitative analysis of protein expression changes. | Employed to compare protein profiles after gene knockdown or temperature stress [99]. |
| Illumina RNA-Seq | High-throughput transcriptome profiling. | Used for comprehensive gene expression analysis across different conditions or life stages [99] [102]. |
The experimental data and methodologies compiled in this guide robustly validate VgR as a critical genetic target for fertility disruption in Z. cucurbitae, with its function being profoundly sensitive to short-term high temperatures. The consistent observation that ZcVgR knockdown leads to a near-total cessation of egg production, coupled with disrupted ovarian development and altered metabolic and immune pathways, provides a compelling case for its central role in reproductive biology. The comparative framework and detailed protocols serve as a foundation for researchers developing RNAi-based biopesticides or exploring thermal biology in pest management.
The health of aquatic ecosystems is increasingly threatened by endocrine-disrupting chemicals (EDCs), a diverse group of contaminants that interfere with hormonal signaling and homeostasis in wildlife [9]. Among the most established tools for detecting estrogenic activity of EDCs is vitellogenin (Vtg), the egg yolk precursor protein normally synthesized by females of oviparous species during oogenesis [47]. In male and juvenile fish, the gene for vitellogenin is present in the liver but remains silent under normal physiological conditions; however, upon exposure to estrogen or estrogen-mimicking chemicals, this gene is activated, leading to the synthesis and secretion of Vtg into the bloodstream [47]. This induction forms the basis of Vtg's use as a sensitive biomarker for detecting estrogenic EDCs in aquatic environments, with applications spanning laboratory research, field monitoring, and regulatory testing [9] [103].
The validation of vitellogenin gene functions is critically important in fertility research, as it provides insights into the molecular mechanisms governing reproductive health. The ecotoxicological application of Vtg induction leverages this fundamental reproductive pathway, using it as a sentinel to detect chemical interference with endocrine function. This review systematically compares the use of Vtg induction across different aquatic models, evaluates experimental protocols for its measurement, and situates these findings within the broader context of reproductive biology and endocrine disruption science.
Vtg Induction Mechanisms: Molecular Pathways and Regulatory Elements
Fundamental Regulatory Pathways
The induction of vitellogenin by estrogenic compounds occurs through a well-characterized molecular pathway. Estrogenic EDCs, acting like endogenous estrogen, bind to and activate estrogen receptors (ERs) in the liver [104]. This receptor-ligand complex then interacts with specific estrogen response elements (EREs) in the promoter region of the Vtg gene, initiating transcription and subsequent Vtg protein synthesis [9] [47]. The synthesized Vtg is then released into the circulatory system, where it can be detected in plasma [104].
Recent multi-omics studies reveal that Vtg induction occurs within broader transcriptional and regulatory networks, involving genes such as cyp19a1 (aromatase), cyp1a (cytochrome P4501A), and other stress-responsive genes [9]. This underscores the complexity of endocrine disruption responses beyond a single biomarker. The molecular regulation of Vtg also shows significant evolutionary diversity; in hemimetabolous insects like Periplaneta americana, for instance, juvenile hormone (JH) rather than ecdysteroids serves as the principal regulator of vitellogenin gene expression [105].
Vtg Gene Diversity and Regulation
Teleost fish exhibit particularly complex Vtg systems, with multiple Vtg genes (e.g., VtgA, VtgB, VtgC) present in many species [103]. These multiple piscine vitellogenins can display different responsiveness to estrogenic compounds, suggesting they may have specialized roles and regulatory mechanisms [103]. The kinetics of induction of distinct Vtg types depends on various environmental factors, including water temperature, photoperiod, life history stage, and the concentration and type of estrogenic compound [103].
Table 1: Key Characteristics of Vitellogenin (Vtg) as an Endocrine Disruption Biomarker
| Characteristic | Description | Significance in Ecotoxicology |
|---|---|---|
| Normal Physiological Expression | Produced by females in response to estradiol during oogenesis | Female-specific reproductive protein |
| Inducible Expression | Can be induced in males, juveniles, and immature females by estrogenic compounds | Sensitive indicator of exposure to estrogenic EDCs |
| Molecular Regulation | Controlled by estrogen receptor binding to estrogen response elements (EREs) in promoter region | Reveals fundamental mechanism of endocrine disruption |
| Protein Detection | Measurable in plasma, serum, and other tissues using immunoassays | Enables quantitative assessment of exposure magnitude |
| Evolutionary Conservation | Found across oviparous vertebrates with conserved functional domains | Allows for cross-species assay development and comparison |
Comparative Analysis of Aquatic Models for Vtg-Based Biomontoring
Fish Models: Established Sentinels
Fish represent the most widely used models for Vtg-based endocrine disruption studies, with several species serving as standard test organisms in international screening programs. The common carp (Cyprinus carpio), fathead minnow (Pimephales promelas), zebrafish (Danio rerio), and Japanese medaka (Oryzias latipes) have been particularly important, with quantitative ELISA methods developed specifically for these species to support standardized testing [106].
The African sharptooth catfish (Clarias gariepinus) has emerged as a particularly valuable model in African freshwater environments, demonstrating detectable plasma Vtg in males from contaminated sites like the Ikpoba and Osse Rivers in Nigeria [104]. This species' nearly pan-African distribution and bottom-dwelling behavior make it especially valuable for understanding contaminant bioavailability in aquatic systems [104]. Field studies have successfully detected plasma Vtg in male C. gariepinus using a sensitive commercially available non-species-specific fish Vtg ELISA, confirming its utility as a biomonitoring species in African freshwater environments [104].
Invertebrate Models: Applications and Limitations
The use of Vtg as a biomarker has extended beyond fish to various invertebrate species, though with notable limitations. In crustaceans like the copepod Eurytemora affinis, Vtg expression shows strong sexual dimorphism, with levels in males ranging from 1,900- to 6,800-fold lower than in females depending on developmental stage [107]. However, unlike in fish models, exposure to known endocrine disruptors like 4-nonylphenol or the crustacean hormone methyl farnesoate did not induce Vtg expression in male copepods [107]. Similarly, studies in mollusks have reported inconsistent induction of Vtg-like proteins, with some finding no induction even under potent EE2 exposure [9]. These findings suggest that Vtg may not be an appropriate biomarker for endocrine disruption across all invertebrate taxa [107].
Table 2: Comparison of Aquatic Models for Vtg-Based Endocrine Disruption Studies
| Species | Model Type | Sensitivity to EDCs | Key Applications | Limitations |
|---|---|---|---|---|
| Fathead minnow (Pimephales promelas) | Standardized test species | High; well-documented Vtg induction | Regulatory testing, laboratory studies | Less relevant for field monitoring in non-native regions |
| Zebrafish (Danio rerio) | Laboratory model | High; used for Vtg ELISA development | Mechanistic studies, developmental toxicology | Not ideal for field assessment |
| African sharptooth catfish (Clarias gariepinus) | Native sentinel species | Demonstrated field detection of Vtg in males | Environmental monitoring in African freshwater systems | Limited commercial assay availability |
| Copepods (Eurytemora affinis) | Invertebrate model | Limited; no significant induction in males shown | Comparative studies in crustaceans | Not reliable as standalone biomarker |
Experimental Protocols and Methodologies
Vtg Detection and Quantification Methods
The detection and quantification of Vtg relies primarily on immunoassay techniques, with enzyme-linked immunosorbent assays (ELISAs) being the most widely used due to their sensitivity, specificity, and capacity for high-throughput analysis [106] [47]. Both sandwich and competitive ELISA formats have been developed, with detection limits typically in the nanogram per milliliter range for plasma samples [106]. For example, carp Vtg ELISA has a working range of 1-63 ng/mL with a minimal detection limit of 0.6 ng/mL, while medaka Vtg ELISA exhibits even greater sensitivity with a working range of 0.25-16 ng/mL and a minimal detection limit of 0.1 ng/mL [106].
The development of species-specific antibodies has been crucial for advancing Vtg quantification. Early approaches focused on generating monoclonal antibodies against purified VTG from model species like rainbow trout (Oncorhynchus mykiss), with selection for cross-reactivity to enable broader application [108]. More recently, polyclonal antisera raised against synthetic consensus peptides representing conserved N-terminal amino acid sequences of Vtg have proven effective for recognizing Vtg across multiple teleost families [108].
Molecular Techniques for Vtg Characterization
Beyond protein detection, molecular methods enable detailed characterization of Vtg gene structure and regulation. Promoter sequencing and analysis have identified critical regulatory elements, such as in Periplaneta americana where a 38 bp region (from -177 to -139 bp) containing two conserved response element half-sites separated by a 2-nucleotide spacer was identified as essential for juvenile hormone III induction of Vg2 [105]. This region, designated Vg2RE, was shown to bind a 71 kDa candidate nuclear protein from previtellogenic female fat body cells [105].
Luciferase reporter assays in cell culture systems (e.g., Sf9 cells) have demonstrated that reporter constructs under the control of minimal promoters containing Vg2RE can be induced by JH III in a dose- and time-dependent manner, providing a powerful tool for dissecting regulatory mechanisms [105]. Electrophoretic mobility shift assays and affinity pull-down experiments further enable the identification of specific protein-DNA interactions involved in Vtg gene regulation [105].
Diagram 1: Molecular Pathway of Vtg Induction by Estrogenic EDCs. This diagram illustrates the key molecular events from EDC exposure to Vtg protein detection in the bloodstream.
Research Reagent Solutions for Vtg Studies
Table 3: Essential Research Reagents for Vtg-Based Endocrine Disruption Studies
| Reagent/Assay Type | Specific Examples | Research Applications | Performance Characteristics |
|---|---|---|---|
| Species-Specific Vtg ELISA | Carp Vtg ELISA, Medaka Vtg ELISA | Quantitative Vtg measurement in standardized test species | Working range: 0.25-63 ng/mL depending on species; intra- and inter-assay variations <20% [106] |
| Cross-Reactive Antibodies | Monoclonal antibodies against conserved Vtg regions | Screening multiple species with single assay platform | Specifically recognizes VTG across phylogenetically distant vertebrates [108] |
| Universal Vtg Assays | Non-species-specific fish Vtg ELISA | Field studies with multiple species or species without dedicated kits | Successfully detected plasma Vtg in wild male C. gariepinus [104] |
| Luciferase Reporter Constructs | Vg promoter-luciferase fusions | Analysis of regulatory elements and transcription factors | Identified Vg2RE as essential for JH III induction in P. americana [105] |
Challenges and Future Directions
Despite the well-established utility of Vtg as a biomarker for endocrine disruption, several challenges remain in its application across different aquatic models. The species-specificity of Vtg molecules often necessitates the development of customized detection assays, which can be resource-intensive [106]. While universal assays offer a promising alternative, their sensitivity may be inferior to species-specific tests [108] [104]. In crustaceans and other invertebrates, the fundamental differences in endocrine systems create limitations for applying Vtg as a biomarker, as evidenced by the lack of inducibility in copepod models [107].
Future directions in Vtg research include the integration of Vtg measurement within Adverse Outcome Pathway (AOP) frameworks and multi-omics approaches that combine transcriptomic, epigenetic, and histological endpoints [9]. The development of sex-specific biomarker panels that incorporate Vtg alongside other molecular indicators holds promise for more comprehensive assessment of endocrine disruption [9]. Advances in ecogenomics and machine learning approaches are further expected to enhance the interpretation of Vtg induction data in environmental monitoring contexts [9].
Diagram 2: Adverse Outcome Pathway (AOP) Framework for EDCs. This diagram places Vtg induction within the broader context of biological organization from molecular initiation to population-level consequences.
Vitellogenin induction remains one of the most sensitive and well-characterized biomarkers for detecting estrogenic endocrine disruption in aquatic environments. While fish models, particularly standardized test species and native sentinels like C. gariepinus, have proven highly effective for Vtg-based monitoring, applications in invertebrate models show significant limitations due to fundamental differences in endocrine regulation. The continued refinement of detection methodologies, including the development of both species-specific and universal assays, enhances our capacity to screen for EDCs across diverse aquatic ecosystems. When integrated within adverse outcome pathway frameworks and multi-omics approaches, Vtg measurement provides a powerful tool for connecting molecular initiation events with higher-level biological effects, ultimately supporting evidence-based environmental management and conservation strategies aimed at preserving aquatic ecosystem health and function.
Comparative Analysis of Vtg Functions in Social Insects vs Solitary Species
Vitellogenin (Vtg), a phospholipoglycoprotein and the precursor to the major egg yolk protein, plays a vital role in the reproduction of oviparous species [109] [110]. Historically regarded as a female-specific reproductive protein, its function has undergone an "evolutionary transformation," with recent research revealing its involvement in non-nutritional roles such as immune defense and antioxidant activity [110]. This protein is of particular significance in the study of social insects, where its functions appear to have been co-opted and expanded beyond ancestral reproductive roles to regulate complex social traits, including division of labor, longevity, and behavior [111] [112] [113]. This guide provides a comparative analysis of Vtg functions in social and solitary insects, framing the discussion within the context of validating gene functions in fertility research. It synthesizes experimental data on Vtg expression, regulation, and evolution, offering methodologies and resources pertinent to researchers and scientists in the field.
The functional profile of vitellogenin differs markedly between solitary and social insects. In solitary insects, Vtg maintains its ancestral role, being primarily dedicated to reproduction [109]. In social insects, however, Vtg has acquired multifunctional characteristics, influencing social organization, caste differentiation, and lifespan [111] [112]. The following table summarizes the core functional differences elucidated from experimental studies.
Table 1: Core Functional Roles of Vitellogenin in Solitary vs. Social Insects
| Functional Aspect | Solitary Insects | Social Insects |
|---|---|---|
| Primary Role | Reproduction; primary energy source for developing embryos [109] [110]. | Reproduction, but also extensively co-opted for social traits [111] [112]. |
| Expression in Females | High in reproductive females; rapid increase after emergence, independent of mating [109]. | Typically highest in reproductive queens; also varies with worker task (e.g., nurses vs. foragers) [112] [113]. |
| Expression in Males | Low levels detected, suggesting potential ancillary functions [109]. | Documented in workers; linked to non-reproductive functions like immunity [109] [112]. |
| Interaction with Juvenile Hormone (JH) | Positive correlation; JH acts as a gonadotropin that induces Vg synthesis [114]. | Highly modified; can be negative (honey bees), uncoupled (bumble bees), or positive depending on lineage [114] [115]. |
| Non-Nutritional Functions | Emerging evidence for antibacterial and antioxidant roles in eggs [110]. | Well-established roles in immunity, antioxidant defense, longevity, and behavioral regulation [111] [110] [112]. |
| Gene Copy Number | Typically a single ortholog [109]. | Frequent gene duplications leading to subfunctionalization (e.g., in ants) [112] [113]. |
A key evolutionary mechanism enabling this functional diversification in social insects is gene duplication. While solitary bees like Centris tarsata and highly eusocial honey bees (Apis mellifera) possess a single Vg ortholog, many ant genomes harbor multiple Vg genes [109] [112] [113]. For instance, the harvester ant Pogonomyrmex barbatus has two Vg genes (Pb_Vg1 and Pb_Vg2), and the fire ant Solenopsis invicta has four [113]. Phylogenetic analyses indicate that an initial duplication event occurred after the divergence of major ant clades, leading to two subfamilies: Subfamily A, which is preferentially expressed in queens and nurses and is associated with reproductive functions, and Subfamily B, which is expressed in foragers and associated with non-reproductive, nutritional functions [113]. This subfunctionalization allows a single ancestral gene to evolve specialized roles, supporting the Reproductive Ground Plan Hypothesis, which proposes that pathways regulating reproduction in solitary ancestors were co-opted to regulate worker division of labor in social insects [113].
Experimental Data and Signaling Pathways
The relationship between Vtg and juvenile hormone (JH) is a pivotal axis that has been rewired during the evolution of sociality, with profound implications for fertility and social organization.
The JH-Vtg Signaling Axis Across Social Contexts
The following diagram illustrates the evolutionary transition of the JH-Vtg relationship from solitary to social insects.
Figure 1: Evolutionary Rewiring of the JH-Vtg Axis. The relationship transitions from a positive correlation in solitary insects to an uncoupled or negative correlation in social species, facilitating the expansion of Vtg's functions.
Quantitative Expression Profiles
Gene expression studies provide concrete data on Vtg's roles. The following table compiles key quantitative findings from representative species.
Table 2: Experimental Vtg Expression Data Across Insect Species
| Species | Caste / Sex | Experimental Condition | Vg Expression Level | Functional Association |
|---|---|---|---|---|
| Solitary Bees (Centris tarsata & C. analis) [109] | Egg-laying Female | Adult life stage | Highest amounts of vg transcripts | Primary role in reproduction |
| Male | Adult life stage | Low, but detectable levels | Ancillary, non-reproductive functions | |
| Bumble Bee (Bombus terrestris) [114] | Fertile Queen | vs. 10-day old Queenless Workers | 6x higher in queens | Association with reproduction |
| Virgin Queen | vs. 4-day old Queenright Workers | 2.4x higher (not significant) | - | |
| Harvester Ant (Pogonomyrmex barbatus) [113] | Queen | vs. Workers | Higher Pb_Vg1 expression | Subfunctionalization; reproductive role |
| Nurse Worker | vs. Foragers | Higher Pb_Vg1 expression | - | |
| Forager Worker | vs. Nurses | Higher Pb_Vg2 expression | Subfunctionalization; non-reproductive role | |
| Black Ant (Formica fusca) [112] | Queen | vs. Workers | Significant upregulation of conventional Vg | Caste differentiation & reproduction |
| Nurse Worker | vs. Foragers | Significant upregulation of conventional Vg | Task specialization & behavior |
Experimental Protocols for Functional Validation
Gene Expression Analysis via qRT-PCR
Objective: To quantify and compare Vg mRNA expression levels across different castes, tissues, or social conditions [114] [109] [112].
- Sample Collection: Dissect target tissues (e.g., fat body, head, whole body for small insects) from individuals of defined caste, age, and social status. Immediately preserve tissue in RNA-later or liquid nitrogen to prevent RNA degradation.
- RNA Extraction: Homogenize tissue and extract total RNA using a commercial kit (e.g., TRIzol reagent or column-based methods). Include a DNase I treatment step to remove genomic DNA contamination.
- cDNA Synthesis: Convert 1 µg of total RNA into complementary DNA (cDNA) using a Reverse Transcription kit with oligo(dT) and/or random hexamer primers.
- Quantitative PCR (qPCR): Prepare reactions with SYBR Green or TaqMan chemistry. Use species-specific primers designed for the Vg gene of interest and a stable reference gene (e.g., actin, rpl32). Run samples in technical triplicates on a real-time PCR machine.
- Data Analysis: Calculate relative gene expression using the comparative Ct (2^(-ΔΔCt)) method. Normalize Vg expression levels to the reference gene and compare statistically between experimental groups using ANOVA or t-tests.
Functional Disruption via RNA Interference (RNAi)
Objective: To investigate the causal relationship between Vg gene activity and social or reproductive phenotypes [111].
- dsRNA Synthesis: Design and synthesize double-stranded RNA (dsRNA) targeting a unique sequence region of the target Vg gene. A gene-specific DNA template is used in an in vitro transcription reaction.
- Delivery of dsRNA: Microinject a defined volume (e.g., 1-2 µL) of dsRNA solution directly into the hemolymph of anesthetized experimental insects (e.g., honey bee workers). A control group should be injected with dsRNA targeting a non-specific gene (e.g., GFP).
- Phenotypic Monitoring: After injection, monitor and record behavioral (e.g., onset of foraging, task preference), physiological (e.g., ovarian activation), and molecular (e.g., validation of Vg knockdown via qRT-PCR) traits.
- Data Interpretation: Compare phenotypes between Vg-knockdown and control groups. A significant change in the measured traits confirms a functional role for Vg in those processes.
The Scientist's Toolkit: Key Research Reagents
Table 3: Essential Reagents for Vitellogenin Research
| Reagent / Solution | Function in Experimental Protocol |
|---|---|
| RNA-later Stabilization Solution | Preserves RNA integrity in tissues post-dissection and during storage [114] [109]. |
| TRIzol Reagent | A monophasic solution of phenol and guanidine isothiocyanate for effective total RNA isolation from insect tissues [113]. |
| DNase I (RNase-free) | Enzymatically degrades genomic DNA during RNA purification to prevent false positives in qRT-PCR [109]. |
| Reverse Transcriptase Kit | Enzyme mix for synthesizing first-strand cDNA from an RNA template, a prerequisite for qPCR [114] [112]. |
| SYBR Green qPCR Master Mix | A fluorescent dye that intercalates with double-stranded DNA, enabling real-time quantification of PCR products [112] [113]. |
| Species-Specific Vg Primers | Short, designed oligonucleotide sequences that flank a unique region of the Vg gene for specific amplification in qPCR [109] [113]. |
| dsRNA for Vg Gene | The effector molecule for RNAi experiments, designed to silence the expression of the target Vg gene [111]. |
| Juvenile Hormone Analogue (e.g., Methoprene) | A synthetic compound used in topical application or injection experiments to manipulate JH titers and study its interaction with Vg [114]. |
This comparative guide underscores that vitellogenin is far more than a passive yolk protein. In solitary insects, it fulfills an essential but primarily reproductive function. In social insects, however, Vtg has undergone a remarkable functional expansion, becoming a key pleiotropic regulator of social organization. This evolutionary process was likely driven by the co-option of ancestral reproductive pathways, a phenomenon strongly supported by the Reproductive Ground Plan Hypothesis. Gene duplication events, particularly prevalent in ants, have further facilitated this process through subfunctionalization, allowing different Vg copies to specialize in reproductive and behavioral tasks. The experimental data and methodologies outlined here provide a framework for researchers to further validate Vtg functions and explore its potential as a model for understanding the evolution of complex traits from simple, ancestral gene networks.
Integrating Histopathological and Molecular Endpoints for Comprehensive Validation
The validation of gene function, particularly for multifaceted genes like vitellogenin (Vg) in fertility research, requires a multi-platform approach that integrates histopathological and molecular endpoints. This guide objectively compares the performance of key methodological platforms—including RNA Interference (RNAi), immunohistochemistry (IHC), and machine learning-based image analysis—for validating vitellogenin's role in fertility. Supporting experimental data from model organisms demonstrates how an integrated strategy overcomes the limitations of single-platform analyses, providing a robust framework for conclusive gene function validation in biomedical research and drug development.
In fertility research, establishing a causal link between a gene's expression and its phenotypic outcome demands more than quantifying mRNA levels. The vitellogenin gene family, essential for yolk formation and embryonic development, exemplifies this complexity. Vgs are not merely biomarkers of fertility but are active regulators of oogenesis and metabolic provisioning [83] [116]. A reductionist approach focusing solely on molecular data may miss critical histopathological manifestations of gene dysfunction, such as impaired oocyte development or ovarian atrophy. Conversely, histology alone may not reveal the underlying molecular mechanisms.
The integration of pathology with molecular biology is vital for enhancing the translational value of biological research [117] [118]. This guide compares the performance of standalone and integrated methods, providing researchers with a clear framework for validating vitellogenin gene functions through a synthesis of histopathological and molecular endpoints.
Comparative Analysis of Methodological Platforms
The table below compares the core methodological platforms used for validating gene functions, highlighting their distinct outputs and performance in providing comprehensive evidence.
Table 1: Performance Comparison of Validation Methodologies for Vitellogenin Gene Function Analysis
| Methodological Platform | Key Outputs and Endpoints | Advantages | Limitations | Suitability for Fertility Validation |
|---|---|---|---|---|
| RNAi/Functional Genetics | Gene knockdown efficiency (qRT-PCR), oocyte count, egg production rate, ovarian morphology (histology) [116] | Estishes causal link; provides quantitative fertility metrics (e.g., egg production) [116] | Off-target effects; may not fully recapitulate chronic or partial loss-of-function | High; directly probes gene function in a physiological context |
| Immunohistochemistry (IHC) | Protein localization and abundance in tissue (e.g., intestine, fat body, ovaries), tissue architecture and morphology [83] [119] | Preserves spatial context; visualizes protein in relevant cell types | Semi-quantitative; dependent on antibody quality and specificity | High; critical for confirming yolk protein production and uptake |
| Transcriptomics (RNA-seq) | Genome-wide differential gene expression, identification of co-expressed genes and pathways [116] | Unbiased discovery; can identify compensatory mechanisms and subtypes | Does not confirm protein presence or function; complex data analysis | Medium; excellent for discovery but requires functional confirmation |
| Machine Learning on Histopathology | Quantitative image features predicting molecular subtypes, mutations, and patient prognosis (e.g., AUC metrics) [120] | Uncovers subtle, prognostically significant patterns invisible to the human eye | "Black box" nature; requires large, well-annotated datasets | Emerging; powerful for linking morphology to molecular features and outcomes |
Detailed Experimental Protocols for Key Assays
Protocol 1: RNAi-Based Functional Validation in an Insect Model
This protocol, adapted from fire ant (Solenopsis invicta) research, details the functional validation of Vg genes [116].
- Objective: To determine the necessity of specific Vg genes (e.g., Vg2, Vg3) for oogenesis and fecundity.
- Experimental Groups: (1) dsRNA targeting Vg2, (2) dsRNA targeting Vg3, (3) dsRNA targeting both, (4) Control group (dsRNA for an unrelated gene, e.g., GFP).
- Procedure:
- dsRNA Synthesis: Design and synthesize double-stranded RNA (dsRNA) targeting specific Vg genes and a control gene.
- Animal Injection: Anesthetize adult female ants/insects and microinject a defined dose of dsRNA (e.g., 500 ng per individual) into the hemolymph.
- Rearing and Monitoring: Maintain insects under standard conditions post-injection.
- Sample Collection: After a set period (e.g., 7-14 days), collect individuals for analysis.
- Endpoint Measurements:
- Molecular Efficacy (qRT-PCR): Isolate RNA from fat bodies or whole abdomens. Use qRT-PCR with gene-specific primers to quantify the knockdown efficiency of target Vg genes relative to the control group.
- Phenotypic Fertility Assessment:
- Ovarian Morphology: Dissect a subset of individuals. Fix ovaries for histology (H&E staining) and score for developmental defects, size, and oocyte maturation.
- Fecundity Quantification: Record the number of eggs laid per female over the experimental period.
- Supporting Data: In S. invicta, knockdown of SiVg2 or SiVg3 resulted in significant downregulation of target mRNA, smaller ovaries, and a drastic reduction in egg production, confirming their critical role in queen fertility [116].
Protocol 2: Integrated Histopathological and Molecular Workflow
This workflow, inspired by hepatocellular carcinoma (HCC) studies, demonstrates how to integrate histopathology with molecular data for a systems-level view [120].
- Objective: To correlate histopathological image features with molecular features (e.g., mutations, gene expression) and clinical outcomes like survival.
- Procedure:
- Tissue Acquisition & Processing: Collect tissue samples (e.g., ovarian or liver tissue). Process for both histology (formalin-fixed, paraffin-embedded blocks) and molecular analysis (snap-freezing for RNA/DNA).
- Histopathological Image Analysis:
- Slide Digitization: Scan whole-slide images at high magnification (e.g., 40x).
- Image Tiling & Feature Extraction: Use software like CellProfiler to tile images and extract hundreds of quantitative features describing cell morphology, texture, and tissue architecture [120].
- Molecular Profiling: In parallel, extract DNA/RNA from corresponding tissue. Perform sequencing or microarrays to identify mutations, gene expression subtypes, or omics profiles.
- Data Integration & Model Building: Use machine learning (e.g., Random Forest, Gradient Boosting) to build predictive models. For example, train a model to predict specific TP53 or CTNNB1 mutations from histopathological image features alone, or to predict patient survival from a combined model of image and omics data [120].
- Endpoint Measurements:
- Model Performance: Assess using Area Under the Curve (AUC) for classification tasks (e.g., mutation prediction) and Concordance Index for survival prediction.
- Clinical Correlation: Validate the model's risk stratification in an independent patient cohort, analyzing survival differences via Kaplan-Meier curves and Cox regression.
- Supporting Data: In HCC, image features predicted TP53 mutations with an AUC of 0.893 and patient 5-year survival with an AUC of 0.819. A multi-platform model combining images and omics achieved a superior 5-year survival AUC of 0.904 [120].
Figure 1: Integrated workflow for combining histopathological and molecular data.
The Scientist's Toolkit: Essential Research Reagents & Solutions
Table 2: Key Research Reagents and Solutions for Vitellogenin and Fertility Studies
| Reagent / Solution | Critical Function in Experimental Protocol | Example Application in Vg Studies |
|---|---|---|
| dsRNA for Target Genes | Induces sequence-specific degradation of mRNA to knock down gene function. | Functional validation of SiVg2 and SiVg3 in red imported fire ants [116]. |
| Vg-Specific Antibodies | Enables detection, localization, and semi-quantification of vitellogenin protein in tissues via IHC. | Confirming Vg protein production in the intestine/fat body and its uptake into oocytes [83]. |
| CellProfiler Software | Open-source platform for automated quantitative analysis of histopathological images. | Extracting features from whole-slide images to predict molecular subtypes [120]. |
| Tissue Microarrays (TMAs) | Allows high-throughput analysis of hundreds of tissue specimens on a single slide. | Validating biomarker expression across large patient cohorts in breast cancer studies [119]. |
| High-Quality RNA/DNA Kits | Isolate intact, pure nucleic acids from frozen or FFPE tissue for downstream molecular assays. | Enabling transcriptomic profiling (RNA-seq) to identify differentially expressed genes [116] [121]. |
| qRT-PCR Reagents | Precisely quantify mRNA expression levels of target genes. | Measuring AlVg transcript abundance in Apolygus lucorum after feeding on different hosts [10]. |
Integrated Signaling and Physiological Pathways
Vitellogenin synthesis and function are central to a complex network that integrates nutrition, hormonal signaling, and reproductive output. The diagram below maps this key pathway.
Figure 2: Vitellogenin pathway integrating environmental cues with reproductive output.
The comparative data and protocols presented demonstrate that no single platform is sufficient for comprehensive validation. The most robust strategy is a multi-platform integration that leverages the strengths of each method.
- For Definitive Functional Validation: RNAi is indispensable for establishing a causal link between Vg gene expression and fertility phenotypes, providing quantitative data on fecundity [116].
- For Spatial Localization and Protein-Level Confirmation: IHC is critical for verifying that Vg protein is synthesized in and transported to the correct tissues [83] [119].
- For Discovery and Unbiased Profiling: Transcriptomics identifies Vg genes and their regulatory networks, especially in non-model organisms [116].
- For Translational Power and Prognostic Insight: Machine learning-based histopathology offers a powerful bridge, quantitatively linking morphological changes to molecular drivers and clinical outcomes, as proven in oncology [120].
Therefore, a tiered approach is recommended: use transcriptomics for discovery, IHC for spatial confirmation, RNAi for functional causality, and integrated computational models to build predictive, clinically relevant frameworks. This multi-faceted methodology ensures that the validation of vitellogenin gene functions—and indeed, any complex gene trait—is both comprehensive and conclusive.
Conclusion
The validation of vitellogenin gene functions establishes it as a master regulator of fertility across diverse species, with remarkable evolutionary conservation in its reproductive roles. Evidence from multiple experimental approaches confirms that Vtg disruption consistently impairs oogenesis, ovarian development, and embryonic viability. The methodological standardization and troubleshooting frameworks developed enable reliable application of Vtg as a sensitive biomarker for endocrine disruption screening and reproductive toxicology assessments. Future research directions should focus on developing sex-specific biomarker panels, integrating omics data with machine learning for predictive modeling, and exploring Vtg's potential as a therapeutic target for fertility management. These advances position Vtg research at the intersection of basic reproductive science, environmental monitoring, and biomedical innovation, offering promising avenues for both conservation biology and clinical applications.
References
- 1. sciencedirect.com/topics/agricultural-and-biological-sciences... [https://www.sciencedirect.com/topics/agricultural-and-biological-sciences/vitellogenin]
- 2. - Wikipedia Vitellogenin [https://en.wikipedia.org/wiki/Vitellogenin]
- 3. The structure of vitellogenin provides a molecular model for the... [https://pubmed.ncbi.nlm.nih.gov/9878414/]
- 4. Vitellogenins - Yolk Gene Function and Regulation in Caenorhabditis elegans - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6736625/]
- 5. Characterization of the low-density lipoprotein receptors family and... [https://www.nature.com/articles/s41598-025-14884-2?error=cookies_not_supported&code=de44e5e8-0528-49cf-ad2e-fbc769770584]
- 6. PAPER TITLE - Study of Vitellogenin | PDF Motif [https://www.slideshare.net/slideshow/paper-title-study-of-vitellogenin-motif/274400559]
- 7. RCSB PDB - 9ENR: Vitellogenin from the honey bee hemolymph [https://www.rcsb.org/structure/9ENR]
- 8. Relationship between reproductive success and male plasma vitellogenin concentrations in cunner, Tautogolabrus adspersus. - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1241311/]
- 9. Sex-Specific Molecular and Genomic Responses to Endocrine Disruptors in Aquatic Species: The Central Role of Vitellogenin [https://www.mdpi.com/2073-4425/16/11/1317]
- 10. Molecular characterisation of the vitellogenin gene (AlVg) and its expression after Apolygus lucorum had fed on different hosts - PubMed [https://pubmed.ncbi.nlm.nih.gov/26663893/]
- 11. New Perspectives on the Evolutionary of Vitellogenin History ... Gene [https://pubmed.ncbi.nlm.nih.gov/30239716/]
- 12. New Perspectives on the Evolutionary History of Vitellogenin Gene Family in Vertebrates - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6185446/]
- 13. Control of Transcriptional Repression of the Vitellogenin Receptor Gene in Largemouth Bass (Micropterus Salmoides) by Select Estrogen Receptors Isotypes - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4271047/]
- 14. Evolution and differential expression of a vertebrate vitellogenin gene cluster | BMC Ecology and Evolution | Full Text [https://bmcecolevol.biomedcentral.com/articles/10.1186/1471-2148-9-2]
- 15. The Gene Has Multiple Coordinating... | PLOS Biology vitellogenin [https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.0050062]
- 16. Differential regulation of hepatopancreatic vitellogenin (VTG) gene expression by two putative molt-inhibiting hormones (MIH1/2) in Pacific white shrimp (Litopenaeus vannamei) - PubMed [https://pubmed.ncbi.nlm.nih.gov/25447412/]
- 17. Ovarian development and role of vitellogenin in reproduction of... gene [https://www.schweizerbart.de/papers/entomologia/detail/44/103030/Ovarian_development_and_role_of_vitellogenin_gene_in_reproduction_of_the_tomato_leaf_miner_Tuta_absoluta]
- 18. Are Cell Junctions Implicated in the Regulation of Vitellogenin Uptake? Insights from an RNAseq-Based Study in Eel, Anguilla australis - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8834532/]
- 19. A bioinformatic approach to characterize the vitellogenin and... receptor [https://www.nature.com/articles/s41598-025-88011-6?error=cookies_not_supported&code=1ca33430-4c7e-405f-b10a-79072ac29c21]
- 20. Molecular Characterization of Vitellogenin and Vitellogenin Receptor of Bemisia tabaci | PLOS One [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0155306]
- 21. Ligand-binding domains in vitellogenin and other... receptors [https://pubmed.ncbi.nlm.nih.gov/9541543/]
- 22. Multivesicular bodies play a key role in vitellogenin by... endocytosis [https://pubmed.ncbi.nlm.nih.gov/2431937/]
- 23. Juvenile Hormone Regulates Vitellogenin Gene Expression through Insulin-like Peptide Signaling Pathway in the Red Flour Beetle, Tribolium castaneum - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3234905/]
- 24. Vitellogenesis - Wikipedia [https://en.wikipedia.org/wiki/Vitellogenesis]
- 25. Molt-inhibiting hormone stimulates vitellogenesis at advanced ovarian... [https://aquaticbiosystems.biomedcentral.com/articles/10.1186/1746-1448-5-7]
- 26. Roles of Vitellogenin and Its Receptor Genes in Female Reproduction... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11857020/]
- 27. Frontiers | Vitellogenins - Yolk Gene and Regulation in... Function [https://www.frontiersin.org/journals/physiology/articles/10.3389/fphys.2019.01067/full]
- 28. Characterization of the low-density lipoprotein receptors family and... [https://pubmed.ncbi.nlm.nih.gov/41022880/]
- 29. Vertebrate Vitellogenin Gene Duplication in Relation to the... | PLOS One [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0000169]
- 30. Duplication, concerted evolution and purifying selection drive the... [https://bmcecolevol.biomedcentral.com/articles/10.1186/1471-2148-10-142]
- 31. Potential regulatory elements of nematode vitellogenin genes revealed by interspecies sequence comparison - PubMed [https://pubmed.ncbi.nlm.nih.gov/2504925/?dopt=Abstract]
- 32. -mediated Double RNAi and Gustatory Perception... Gene Knockdown [https://www.jove.com/t/50446/rnai-mediated-double-gene-knockdown-gustatory-perception-measurement]
- 33. to RNA Expression Interference Knock Down Gene [https://pubmed.ncbi.nlm.nih.gov/29423805/]
- 34. A direct comparison of strategies for combinatorial RNA interference [https://bmcmolbiol.biomedcentral.com/articles/10.1186/1471-2199-11-77]
- 35. Virus-induced gene silencing for in planta validation of gene ... function [https://pubmed.ncbi.nlm.nih.gov/35944218/]
- 36. Brain gene expression changes elicited by peripheral vitellogenin knockdown in the honey bee - PubMed [https://pubmed.ncbi.nlm.nih.gov/23889463/]
- 37. Vitellogenin family gene expression does not... | F1000 Research [https://f1000research.com/articles/3-125]
- 38. RNA from a simple-tandem repeat is required for sperm maturation and male fertility in Drosophila melanogaster | eLife [https://elifesciences.org/articles/48940]
- 39. An interactive web-based application for Comprehensive Analysis of RNAi-screen Data | Nature Communications [https://www.nature.com/articles/ncomms10578]
- 40. Reverse transcription polymerase chain reaction - Wikipedia [https://en.wikipedia.org/wiki/Reverse_transcription_polymerase_chain_reaction]
- 41. Monitoring gene expression: quantitative real-time rt-PCR - PubMed [https://pubmed.ncbi.nlm.nih.gov/23912981/]
- 42. Artificial intelligence in in-vitro fertilization (IVF): A new era of precision and personalization in fertility treatments - ScienceDirect [https://www.sciencedirect.com/science/article/pii/S246878472400182X]
- 43. Choice Of One Step RT-qPCR Or Two Step RT-qPCR | NEB [https://www.neb.com/en-us/applications/dna-amplification-pcr-and-qpcr/choice-of-one-step-rt-qpcr-or-two-step-rt-qpcr]
- 44. of gene Comparison by reverse transcription... expression profiling [https://pubmed.ncbi.nlm.nih.gov/20569212/]
- 45. Reproducibility of quantitative RT- PCR array in miRNA expression ... [https://bmcgenomics.biomedcentral.com/articles/10.1186/1471-2164-10-407]
- 46. Experimental comparison of relative RT-qPCR quantification approaches for gene expression studies in poplar - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC2930637/]
- 47. Vitellogenin as a Biomarker of Exposure for Estrogen or Estrogen Mimics | Ecotoxicology [https://link.springer.com/article/10.1023/A:1008986522208]
- 48. Experimental evaluation of vitellogenin as a predictive biomarker for reproductive disruption - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC1240371/]
- 49. Decoding Vitellogenin Subtype Responses: A Molecular Approach to Biomarkers of Endocrine Disruption in Scatophagus argus [https://www.mdpi.com/2410-3888/10/1/15]
- 50. Determination of mRNA expression of DMRT93B, vitellogenin , and... [https://pubmed.ncbi.nlm.nih.gov/21656159/]
- 51. Vitellogenin – a biomarker for endocrine disruptors - ScienceDirect [https://www.sciencedirect.com/science/article/abs/pii/S016599369800020X]
- 52. Single-cell multi - omics : Integrating the transcriptome... | Abcam [https://www.abcam.com/en-us/stories/articles/integrating-the-transcriptome-epigenome-and-proteome]
- 53. Top Single Cell Multi - Omics Companies & How to Compare Them... [https://www.linkedin.com/pulse/top-single-cell-multi-omics-companies-how-nks5e]
- 54. Epigenetics analysis - Latest research and news | Nature [https://www.nature.com/subjects/epigenetics-analysis]
- 55. New Generation of Clinical Epigenetics Analysis and Diagnosis for Precision Medicine - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12192280/]
- 56. Transcriptomics technologies - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC5436640/]
- 57. High-throughput epigenetic profiling immunoassays for accelerated disease research and clinical development - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12269834/]
- 58. Network-based multi - omics integrative analysis methods in drug... [https://biodatamining.biomedcentral.com/articles/10.1186/s13040-025-00442-z]
- 59. Adaptation of High - Throughput in Screening ... Drug [https://pmc.ncbi.nlm.nih.gov/articles/PMC3269696/]
- 60. - High in Throughput ... | Technology Networks Screening Drug [https://www.technologynetworks.com/drug-discovery/articles/high-throughput-screening-principles-applications-and-advancements-405121]
- 61. Comprehensive and unbiased multiparameter high - throughput ... [https://elifesciences.org/articles/73760]
- 62. Market | Global Market Analysis Report... High Throughput Screening [https://www.futuremarketinsights.com/reports/high-throughput-screening-market]
- 63. Highthroughput Ecotoxicology (HITEC) - Helmholtz-Centre for... [https://www.ufz.de/index.php?en=50906]
- 64. Downloadable Computational Toxicology Data | US EPA [https://www.epa.gov/comptox-tools/downloadable-computational-toxicology-data]
- 65. Genomic disturbance of vitellogenin 2 (vtg2) leads to vitellin membrane deficiencies and significant mortalities at early stages of embryonic development in zebrafish (Danio rerio) | Scientific Reports [https://www.nature.com/articles/s41598-023-46148-2]
- 66. Tools to minimize interlaboratory in variability gene... vitellogenin [https://pubmed.ncbi.nlm.nih.gov/28631833/]
- 67. and associated Quantification of induced variability ... vitellogenin [https://academic.oup.com/etc/article-abstract/26/2/287/7763461?redirectedFrom=fulltext]
- 68. Molecular characterization and CRISPR/Cas9 validation of the... [https://pubmed.ncbi.nlm.nih.gov/39277149/]
- 69. RNA interference - Wikipedia [https://en.wikipedia.org/wiki/RNA_interference]
- 70. Optimizing siRNA Transfection | Thermo Fisher Scientific - US [https://www.thermofisher.com/us/en/home/references/gibco-cell-culture-basics/transfection-basics/guidelines-for-rna-transfection/optimizing-sirna-transfection.html]
- 71. Optimizing siRNA Transfection for RNAi | Thermo Fisher Scientific - US [https://www.thermofisher.com/us/en/home/references/ambion-tech-support/rnai-sirna/tech-notes/optimizing-sirna-transfection-for-rnai.html]
- 72. CRISPR Delivery Methods: Cargo, Vehicles, and Challenges [https://www.synthego.com/blog/delivery-crispr-cas9/]
- 73. Nanotechnology-driven gene silencing: advancements in SIGS–dsRNA technology for sustainable disease management | Chemical and Biological Technologies in Agriculture | Full Text [https://chembioagro.springeropen.com/articles/10.1186/s40538-025-00738-6]
- 74. sequences for RNAi in pest control and research... Optimizing dsRNA [https://bmcbiol.biomedcentral.com/articles/10.1186/s12915-025-02219-6]
- 75. Disruption of vitellogenin gene function in adult honeybees by intra-abdominal injection of double-stranded RNA - PubMed [https://pubmed.ncbi.nlm.nih.gov/12546706/]
- 76. Genotype effect on regulation of behaviour by vitellogenin supports reproductive origin of honeybee foraging bias - PubMed [https://pubmed.ncbi.nlm.nih.gov/20454635/]
- 77. Genome-Wide Identification of Vitellogenin Gene Family and Comparative Analysis of Their Involvement in Ovarian Maturation in Exopalaemon carinicauda - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC10815947/]
- 78. in Honey Bee Behavior and Lifespan | SpringerLink Vitellogenin [https://link.springer.com/chapter/10.1007/978-94-007-2099-2_2]
- 79. Sensitive and robust gene expression changes in fish exposed to... [https://bmcgenomics.biomedcentral.com/articles/10.1186/1471-2164-8-149]
- 80. RNA integrity and the effect on the real-time qRT-PCR performance - PubMed [https://pubmed.ncbi.nlm.nih.gov/16469371/]
- 81. of relative mRNA quantification models and the impact of... Comparison [https://pubmed.ncbi.nlm.nih.gov/16900335/]
- 82. A PCR-based quantitative assay for the evaluation of mRNA integrity in rat samples - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6006387/]
- 83. Frontiers | Vitellogenins - Yolk Gene Function and Regulation in ... [https://www.frontiersin.org/articles/10.3389/fphys.2019.01067/full]
- 84. quantification and quality assessment techniques | QIAGEN RNA [https://www.qiagen.com/us/knowledge-and-support/knowledge-hub/bench-guide/rna/rna-analysis/quantification-and-analysis-of-rna]
- 85. PCR efficiency [https://www.qiagen.com/us/knowledge-and-support/knowledge-hub/bench-guide/pcr/pcr-quantification/determining-amplification-efficiencies]
- 86. Assay Optimization and PCR Validation [https://www.sigmaaldrich.com/GU/en/technical-documents/technical-article/genomics/pcr/assay-optimization-and-validation]
- 87. Per-channel basis normalization methods for flow cytometry data - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3648208/]
- 88. High Throughput Flow Cytometry Data Normalization for Clinical Trials - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3992339/]
- 89. Unlocking Fluorescence Detector Secrets [https://www.numberanalytics.com/blog/ultimate-guide-fluorescence-detector]
- 90. How to Interpret Flow Cytometry Data | Fortis Life Sciences [https://www.fortislife.com/resources/antibody-resources/how-to-interpret-flow-cytometry-data]
- 91. Comparative Analyses of Reproductive Caste Types Reveal... [https://pubmed.ncbi.nlm.nih.gov/38138959/]
- 92. Flow cytometry multi-color selector [https://www.abcam.com/flow-cytometry/]
- 93. (RHYCFE)[Datasheet]| EPPO Global... Rhynchophorus ferrugineus [https://gd.eppo.int/taxon/RHYCFE/datasheet]
- 94. Current Status of Biology–Biotechnic, Agronomic, and Biological Control of Rhynchophorus ferrugineus: A Review [https://www.mdpi.com/2075-4450/15/12/955]
- 95. Silencing of vitellogenin gene contributes to the promise of controlling... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8568968/]
- 96. -mediated silencing of RNAi curtails oogenesis in the... vitellogenin gene [https://pmc.ncbi.nlm.nih.gov/articles/PMC7877660/]
- 97. Insecticidal potency of RNAi-based catalase knockdown in Rhynchophorus ferrugineus (Oliver) (Coleoptera: Curculionidae) - PubMed [https://pubmed.ncbi.nlm.nih.gov/26822903/]
- 98. Frontiers | Function of Vitellogenin receptor gene in reproductive... [https://www.frontiersin.org/journals/physiology/articles/10.3389/fphys.2022.995004/full]
- 99. Knockdown of the ZcVgR Gene Alters the Expression of Genes... [https://pubmed.ncbi.nlm.nih.gov/39689075/]
- 100. The ZcVg3 Gene Regulates the Reproduction and Lifespan of Female... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11277402/]
- 101. Multigenerational Effects of Short-Term High Temperature on the... [https://www.mdpi.com/2077-0472/12/7/954]
- 102. Genome-wide gene expression profiling of the melon fly, Zeugodacus cucurbitae, during thirteen life stages | Scientific Data [https://www.nature.com/articles/s41597-020-0387-9]
- 103. [PDF] Multiple piscine vitellogenins: biomarkers of... | Semantic Scholar [https://www.semanticscholar.org/paper/Multiple-piscine-vitellogenins%3A-biomarkers-of-fish-Hiramatsu-Matsubara/d56fa679ac2ef2ac61b628c1b467b3334bfabbac]
- 104. Frontiers | Plasma vitellogenin detection in males of the African... [https://www.frontiersin.org/journals/environmental-science/articles/10.3389/fenvs.2025.1622837/full]
- 105. Frontiers | Involvement of Cis-Acting Elements in Molecular Regulation... [https://www.frontiersin.org/journals/physiology/articles/10.3389/fphys.2021.723072/full]
- 106. Development of quantitative vitellogenin-ELISAs for fish test species used in endocrine disruptor screening - PubMed [https://pubmed.ncbi.nlm.nih.gov/14551668/]
- 107. Controversial use of vitellogenin as a biomarker of endocrine ... [https://pubmed.ncbi.nlm.nih.gov/28974407/]
- 108. Universal assay of vitellogenin as a biomarker for environmental estrogens - PubMed [https://pubmed.ncbi.nlm.nih.gov/8593883/]
- 109. Vitellogenin of the solitary bees Centris tarsata and Centris analis (Hymenoptera: Apidae): cDNA structural analysis and gene expression | Apidologie [https://link.springer.com/article/10.1007/s13592-020-00818-6]
- 110. Functions of Vitellogenin in Eggs - PubMed [https://pubmed.ncbi.nlm.nih.gov/28779327/]
- 111. The gene vitellogenin has multiple coordinating effects on social ... [https://pubmed.ncbi.nlm.nih.gov/17341131/]
- 112. and Vitellogenin -like gene expression patterns in relation... vitellogenin [https://link.springer.com/article/10.1007/s00040-019-00725-9]
- 113. Vitellogenin Underwent Subfunctionalization to Acquire Caste and Behavioral Specific Expression in the Harvester Ant Pogonomyrmex barbatus | PLOS Genetics [https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1003730]
- 114. Exploring the role of juvenile hormone and vitellogenin in reproduction... [https://bmcecolevol.biomedcentral.com/articles/10.1186/1471-2148-14-45]
- 115. Transcriptomic Signatures of Ageing Vary in Solitary and Social ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8214409/]
- 116. Comparative Analyses of Reproductive Caste Types Reveal Vitellogenin Genes Involved in Queen Fertility in Solenopsis invicta [https://www.mdpi.com/1422-0067/24/24/17130]
- 117. Why is it crucial to reintegrate pathology into cancer research? - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6377259/]
- 118. Why is it crucial to reintegrate pathology into cancer research? - PubMed [https://pubmed.ncbi.nlm.nih.gov/21590787/]
- 119. subtypes, Molecular grade and survival in a historic... histopathological [https://link.springer.com/article/10.1007/s10549-013-2647-2]
- 120. Frontiers | Multi-platform integration of histopathological images and... [https://www.frontiersin.org/journals/oncology/articles/10.3389/fonc.2025.1591165/full]
- 121. Molecular Pathology | Herbert Irving Comprehensive Cancer Center (HICCC) - New York [https://www.cancer.columbia.edu/research/shared-resources/molecular-pathology]